Vinyl-butyrate
General
Type : Vinyl alkyl ester || Alkyl ester || Butyrate || Water-soluble short chain ester
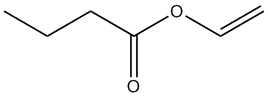
Chemical_Nomenclature : ethenyl butanoate
Canonical SMILES : CCCC(=O)OC=C
InChI : InChI=1S\/C6H10O2\/c1-3-5-6(7)8-4-2\/h4H,2-3,5H2,1H3
InChIKey : MEGHWIAOTJPCHQ-UHFFFAOYSA-N
Other name(s) : Butanoic acid, ethenyl ester, Vinyl butanoate, ethenyl butanoate, Butyric acid, vinyl ester, NSC 5280, Vinylester kyseliny maselne [Czech], EINECS 204-609-4, UN2838, BRN 1744933, AI3-24888, Butyric Acid Vinyl Ester, Vinyl butyrate, inhibited, Vinylester kyseliny maselne, butanoic acid ethenyl ester, AC1L1L8A, 19390_ALDRICH, 19390_FLUKA, NSC5280, MolPort-000-159-328, NSC-5280, Butanoic acid, ethenyl ester (9CI), ZINC00404766, AKOS006223438, LS-48208, B0765, 15560-EP2305683A1, 15560-EP2311839A1, 15560-EP2314589A1, 15560-EP2316837A1, 15560-EP2380661A2, A805027, I14-54174, Vinyl butyrate, inhibited [UN2838] [Flammable liquid], InChI=1\/C6H10O2\/c1-3-5-6(7)8-4-2\/h4H,2-3,5H2,1H
Target
Families : Acidic_Lipase, Cholesterol_esterase, Carb_B_Chordata
References (3)
| Title : Non-lipolytic and lipolytic sequence-related carboxylesterases: a comparative study of the structure-function relationships of rabbit liver esterase 1 and bovine pancreatic bile-salt-activated lipase - Chahinian_2010_Biochim.Biophys.Acta_1801_1195 |
| Author(s) : Chahinian H , Fantini J , Garmy N , Manco G , Sarda L |
| Ref : Biochimica & Biophysica Acta , 1801 :1195 , 2010 |
| Abstract : Chahinian_2010_Biochim.Biophys.Acta_1801_1195 |
| ESTHER : Chahinian_2010_Biochim.Biophys.Acta_1801_1195 |
| PubMedSearch : Chahinian_2010_Biochim.Biophys.Acta_1801_1195 |
| PubMedID: 20655391 |
| Title : How gastric lipase, an interfacial enzyme with a Ser-His-Asp catalytic triad, acts optimally at acidic pH - Chahinian_2006_Biochemistry_45_993 |
| Author(s) : Chahinian H , Snabe T , Attias C , Fojan P , Petersen SB , Carriere F |
| Ref : Biochemistry , 45 :993 , 2006 |
| Abstract : Chahinian_2006_Biochemistry_45_993 |
| ESTHER : Chahinian_2006_Biochemistry_45_993 |
| PubMedSearch : Chahinian_2006_Biochemistry_45_993 |
| PubMedID: 16411775 |