Polyurethane
General
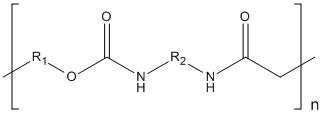
Type : Polymer || Carbamate || Polyurethane
Chemical_Nomenclature :
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : CHEBI:60737, PUR
MW :
Formula :
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
References (14)
| Title : Identification and characterization of a fungal cutinase-like enzyme CpCut1 from Cladosporium sp. P7 for polyurethane degradation - Liu_2024_Appl.Environ.Microbiol__e0147723 |
| Author(s) : Liu J , Xin K , Zhang T , Wen Y , Li D , Wei R , Zhou J , Cui Z , Dong W , Jiang M |
| Ref : Applied Environmental Microbiology , :e0147723 , 2024 |
| Abstract : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| ESTHER : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| PubMedSearch : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| PubMedID: 38445906 |
| Gene_locus related to this paper: 9pezi-CpCut1 |
| Title : Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database-PAZy - Buchholz_2022_Proteins_90_1443 |
| Author(s) : Buchholz PCF , Feuerriegel G , Zhang H , Perez-Garcia P , Nover LL , Chow J , Streit WR , Pleiss J |
| Ref : Proteins , 90 :1443 , 2022 |
| Abstract : Buchholz_2022_Proteins_90_1443 |
| ESTHER : Buchholz_2022_Proteins_90_1443 |
| PubMedSearch : Buchholz_2022_Proteins_90_1443 |
| PubMedID: 35175626 |
| Title : Cloning, expression and characterization of PURase gene from Pseudomonas sp. AKS31 - Saha_2022_Arch.Microbiol_204_498 |
| Author(s) : Saha M , Dutta SP , Mukherjee G , Basu A , Majumder D , Sil AK |
| Ref : Arch Microbiol , 204 :498 , 2022 |
| Abstract : Saha_2022_Arch.Microbiol_204_498 |
| ESTHER : Saha_2022_Arch.Microbiol_204_498 |
| PubMedSearch : Saha_2022_Arch.Microbiol_204_498 |
| PubMedID: 35849211 |
| Gene_locus related to this paper: psesp-PURase |
| Title : Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects - Liu_2021_Biotechnol.Adv_48_107730 |
| Author(s) : Liu J , He J , Xue R , Xu B , Qian X , Xin F , Blank LM , Zhou J , Wei R , Dong W , Jiang M |
| Ref : Biotechnol Adv , 48 :107730 , 2021 |
| Abstract : Liu_2021_Biotechnol.Adv_48_107730 |
| ESTHER : Liu_2021_Biotechnol.Adv_48_107730 |
| PubMedSearch : Liu_2021_Biotechnol.Adv_48_107730 |
| PubMedID: 33713745 |
| Title : Polyurethanases: Three-dimensional structures and molecular dynamics simulations of enzymes that degrade polyurethane - do Canto_2019_J.Mol.Graph.Model_89_82 |
| Author(s) : do Canto VP , Thompson CE , Netz PA |
| Ref : J Mol Graph Model , 89 :82 , 2019 |
| Abstract : do Canto_2019_J.Mol.Graph.Model_89_82 |
| ESTHER : do Canto_2019_J.Mol.Graph.Model_89_82 |
| PubMedSearch : do Canto_2019_J.Mol.Graph.Model_89_82 |
| PubMedID: 30877946 |
| Gene_locus related to this paper: psech-PUEA , psech-PUEB , psefl-PULA |
| Title : Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation - Danso_2019_Appl.Environ.Microbiol_85_e01095 |
| Author(s) : Danso D , Chow J , Streit WR |
| Ref : Applied Environmental Microbiology , 85 : , 2019 |
| Abstract : Danso_2019_Appl.Environ.Microbiol_85_e01095 |
| ESTHER : Danso_2019_Appl.Environ.Microbiol_85_e01095 |
| PubMedSearch : Danso_2019_Appl.Environ.Microbiol_85_e01095 |
| PubMedID: 31324632 |
| Title : Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks - Magnin_2019_Waste.Manag_85_141 |
| Author(s) : Magnin A , Pollet E , Perrin R , Ullmann C , Persillon C , Phalip V , Averous L |
| Ref : Waste Manag , 85 :141 , 2019 |
| Abstract : Magnin_2019_Waste.Manag_85_141 |
| ESTHER : Magnin_2019_Waste.Manag_85_141 |
| PubMedSearch : Magnin_2019_Waste.Manag_85_141 |
| PubMedID: 30803567 |
| Title : PMBD: a Comprehensive Plastics Microbial Biodegradation Database - Gan_2019_Database.(Oxford)_2019_bav119 |
| Author(s) : Gan Z , Zhang H |
| Ref : Database (Oxford) , 2019 :bav119 , 2019 |
| Abstract : Gan_2019_Database.(Oxford)_2019_bav119 |
| ESTHER : Gan_2019_Database.(Oxford)_2019_bav119 |
| PubMedSearch : Gan_2019_Database.(Oxford)_2019_bav119 |
| PubMedID: 31738435 |
| Title : Degradation of Polyester Polyurethane by Bacterial Polyester Hydrolases - Schmidt_2017_Polymers.(Basel)_9_65 |
| Author(s) : Schmidt J , Wei R , Oeser T , Dedavid e Silva LA , Breite D , Schulze A , Zimmermann W |
| Ref : Polymers (Basel) , 9 :65 , 2017 |
| Abstract : Schmidt_2017_Polymers.(Basel)_9_65 |
| ESTHER : Schmidt_2017_Polymers.(Basel)_9_65 |
| PubMedSearch : Schmidt_2017_Polymers.(Basel)_9_65 |
| PubMedID: 30970745 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 , 9bact-g9by57 , thefu-q6a0i4 , thefu-q6a0i3 |
| Title : Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? - Wei_2017_Microb.Biotechnol_10_1308 |
| Author(s) : Wei R , Zimmermann W |
| Ref : Microb Biotechnol , 10 :1308 , 2017 |
| Abstract : Wei_2017_Microb.Biotechnol_10_1308 |
| ESTHER : Wei_2017_Microb.Biotechnol_10_1308 |
| PubMedSearch : Wei_2017_Microb.Biotechnol_10_1308 |
| PubMedID: 28371373 |
| Title : Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata - Wei_2014_AMB.Express_4_44 |
| Author(s) : Wei R , Oeser T , Then J , Kuhn N , Barth M , Schmidt J , Zimmermann W |
| Ref : AMB Express , 4 :44 , 2014 |
| Abstract : Wei_2014_AMB.Express_4_44 |
| ESTHER : Wei_2014_AMB.Express_4_44 |
| PubMedSearch : Wei_2014_AMB.Express_4_44 |
| PubMedID: 25405080 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 |
| Title : Biodegradation of polyester polyurethane by endophytic fungi - Russell_2011_Appl.Environ.Microbiol_77_6076 |
| Author(s) : Russell JR , Huang J , Anand P , Kucera K , Sandoval AG , Dantzler KW , Hickman D , Jee J , Kimovec FM , Koppstein D , Marks DH , Mittermiller PA , Nunez SJ , Santiago M , Townes MA , Vishnevetsky M , Williams NE , Vargas MP , Boulanger LA , Bascom-Slack C , Strobel SA |
| Ref : Applied Environmental Microbiology , 77 :6076 , 2011 |
| Abstract : Russell_2011_Appl.Environ.Microbiol_77_6076 |
| ESTHER : Russell_2011_Appl.Environ.Microbiol_77_6076 |
| PubMedSearch : Russell_2011_Appl.Environ.Microbiol_77_6076 |
| PubMedID: 21764951 |
| Title : Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium - Gautam_2007_Biotechnol.Lett_29_1081 |
| Author(s) : Gautam R , Bassi AS , Yanful EK |
| Ref : Biotechnol Lett , 29 :1081 , 2007 |
| Abstract : Gautam_2007_Biotechnol.Lett_29_1081 |
| ESTHER : Gautam_2007_Biotechnol.Lett_29_1081 |
| PubMedSearch : Gautam_2007_Biotechnol.Lett_29_1081 |
| PubMedID: 17450322 |
| Title : Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB-35 - Nomura_1998_J.Ferment.Bioeng_86_339 |
| Author(s) : Nomura N , Shigeno-Akutsu Y , Nakajima-kambe T , Nakahara T |
| Ref : J.Ferment.Bioeng , 86 :339 , 1998 |
| Abstract : Nomura_1998_J.Ferment.Bioeng_86_339 |
| ESTHER : Nomura_1998_J.Ferment.Bioeng_86_339 |
| PubMedSearch : Nomura_1998_J.Ferment.Bioeng_86_339 |
| PubMedID: |
| Gene_locus related to this paper: comac-polest |