Phaeophytin-a
General
Type : Natural || Chlorophyll-like
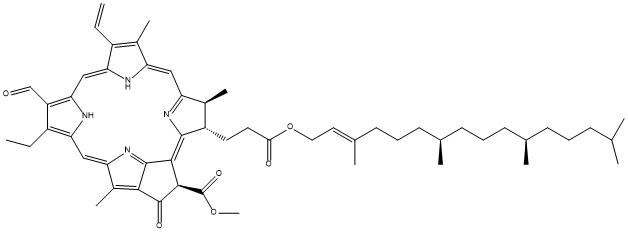
Chemical_Nomenclature : methyl (3R,21S,22S)-16-ethenyl-11-ethyl-4-hydroxy-12,17,21,26-tetramethyl-22-[3-oxo-3-[(E,7R,11R)-3,7,11,15-tetramethylhexadec-2-enoxy]propyl]-7,23,24,25-tetrazahexacyclo[18.2.1.15,8.110,13.115,18.02,6]hexacosa-1,4,6,8(26),9,11,13(25),14,16,18(24),19-undecaene-3-carboxylate
Canonical SMILES : CCC1=C(C2=NC1=CC3=C(C4=C(C(C(=C5C(C(C(=CC6=NC(=C2)C(=C6C)C=C)N5)C)CCC(=O)OCC=C(C)CCCC(C)CCCC(C)CCCC(C)C)C4=N3)C(=O)OC)O)C)C
InChI : InChI=1S\/C55H74N4O5\/c1-13-39-35(8)42-28-44-37(10)41(24-25-48(60)64-27-26-34(7)23-17-22-33(6)21-16-20-32(5)19-15-18-31(3)4)52(58-44)50-51(55(62)63-12)54(61)49-38(11)45(59-53(49)50)30-47-40(14-2)36(9)43(57-47)29-46(39)56-42\/h13,26,28-33,37,41,51,58,61H,1,14-25,27H2,2-12H3\/b34-26+,44-28?,46-29?,47-30?,52-50?\/t32-,33-,37+,41+,51-\/m1\/s1
InChIKey : FDHFJXKRMIVNCQ-OSIZZBRKSA-N
Other name(s) : Pheophytin a, PLU1CG1U91, CHEBI:44898, 3-Phorbinepropanoic acid, 9-ethenyl-14-ethyl-21-(methoxycarbonyl)-4,8,13,18-tetramethyl-20-oxo-, (2E,7R,11R)-3,7,11,15-tetramethyl-2-hexadecenyl ester, (3S,4S,21R)-
Target
Families : Pheophytinase, Chlorophyllase_Plant, Chlorophyllase
References (4)
| Title : A structure-function analysis of chlorophyllase reveals a mechanism for activity regulation dependent on disulfide bonds - Jo_2023_J.Biol.Chem__102958 |
| Author(s) : Jo M , Knapp M , Boggs DG , Brimberry M , Donnan PH , Bridwell-Rabb J |
| Ref : Journal of Biological Chemistry , :102958 , 2023 |
| Abstract : Jo_2023_J.Biol.Chem__102958 |
| ESTHER : Jo_2023_J.Biol.Chem__102958 |
| PubMedSearch : Jo_2023_J.Biol.Chem__102958 |
| PubMedID: 36731794 |
| Gene_locus related to this paper: wheat-w5h2c8 |
| Title : Molecular, structural and biochemical characterization of a novel recombinant chlorophyllase from cyanobacterium Oscillatoria acuminata PCC 6304 - Gu_2021_Microb.Cell.Fact_20_14 |
| Author(s) : Gu S , Dai X , Xu Z , Niu Q , Jiang J , Liu Y |
| Ref : Microb Cell Fact , 20 :14 , 2021 |
| Abstract : Gu_2021_Microb.Cell.Fact_20_14 |
| ESTHER : Gu_2021_Microb.Cell.Fact_20_14 |
| PubMedSearch : Gu_2021_Microb.Cell.Fact_20_14 |
| PubMedID: 33430874 |
| Title : A Novel Recombinant Chlorophyllase1 from Chlamydomonas reinhardtii for the Production of Chlorophyllide Derivatives - Chou_2015_J.Agric.Food.Chem_63_9496 |
| Author(s) : Chou YL , Ko CY , Yen CC , Chen LF , Shaw JF |
| Ref : Journal of Agricultural and Food Chemistry , 63 :9496 , 2015 |
| Abstract : Chou_2015_J.Agric.Food.Chem_63_9496 |
| ESTHER : Chou_2015_J.Agric.Food.Chem_63_9496 |
| PubMedSearch : Chou_2015_J.Agric.Food.Chem_63_9496 |
| PubMedID: 26478543 |
| Gene_locus related to this paper: chlre-a8j2s9 |
| Title : Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis - Schelbert_2009_Plant.Cell_21_767 |
| Author(s) : Schelbert S , Aubry S , Burla B , Agne B , Kessler F , Krupinska K , Hortensteiner S |
| Ref : Plant Cell , 21 :767 , 2009 |
| Abstract : Schelbert_2009_Plant.Cell_21_767 |
| ESTHER : Schelbert_2009_Plant.Cell_21_767 |
| PubMedSearch : Schelbert_2009_Plant.Cell_21_767 |
| PubMedID: 19304936 |
| Gene_locus related to this paper: arath-Q9FFZ1 |