Paranitrophenyl-palmitate
General
Type : pNP || Chromogen || Palmitate || Hexadecanoate || Water-insoluble medium and long chain ester
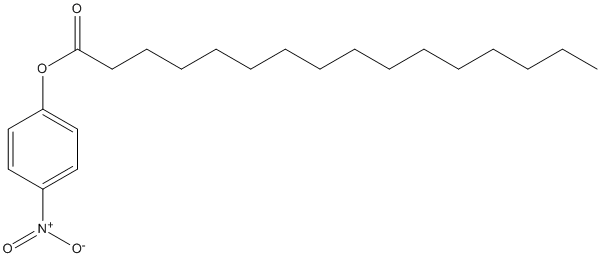
Chemical_Nomenclature : (4-nitrophenyl) hexadecanoate
Canonical SMILES : CCCCCCCCCCCCCCCC(=O)OC1=CC=C(C=C1)[N+](=O)[O-]
InChI : InChI=1S\/C22H35NO4\/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-22(24)27-21-18-16-20(17-19-21)23(25)26\/h16-19H,2-15H2,1H3
InChIKey : LVZSQWIWCANHPF-UHFFFAOYSA-N
Other name(s) : pNP-C16, 4-Nitrophenyl palmitate, P-Nitrophenyl palmitate, p-NP palmitate, pNP-palmitate, 4-NPP, p-NPP, pNPP, 4-nitrophenyl hexadecanoate, Hexadecanoic acid 4-nitrophenyl ester, (4-nitrophenyl) hexadecanoate
References (5)
| Title : Heterologous expression, purification, and characterization of a recombinant Cordyceps militaris lipase from Candida rugosa-like family in Pichia pastoris - Lee_2023_Enzyme.Microb.Technol_168_110254 |
| Author(s) : Lee J , Lee H , Chang PS |
| Ref : Enzyme Microb Technol , 168 :110254 , 2023 |
| Abstract : Lee_2023_Enzyme.Microb.Technol_168_110254 |
| ESTHER : Lee_2023_Enzyme.Microb.Technol_168_110254 |
| PubMedSearch : Lee_2023_Enzyme.Microb.Technol_168_110254 |
| PubMedID: 37201411 |
| Gene_locus related to this paper: cormm-g3j8e9 |
| Title : A Novel Lipase from Streptomyces exfoliatus DSMZ 41693 for Biotechnological Applications - Rodriguez-Alonso_2023_Int.J.Mol.Sci_24_17071 |
| Author(s) : Rodriguez-Alonso G , Toledo-Marcos J , Serrano-Aguirre L , Rumayor C , Pasero B , Flores A , Saborido A , Hoyos P , Hernaiz MJ , de la Mata I , Arroyo M |
| Ref : Int J Mol Sci , 24 : , 2023 |
| Abstract : Rodriguez-Alonso_2023_Int.J.Mol.Sci_24_17071 |
| ESTHER : Rodriguez-Alonso_2023_Int.J.Mol.Sci_24_17071 |
| PubMedSearch : Rodriguez-Alonso_2023_Int.J.Mol.Sci_24_17071 |
| PubMedID: 38069394 |
| Gene_locus related to this paper: strex-SeLipC |
| Title : Phylogenetic analysis and in-depth characterization of functionally and structurally diverse CE5 cutinases - Novy_2021_J.Biol.Chem__101302 |
| Author(s) : Novy V , Carneiro LV , Shin JH , Larsbrink J , Olsson L |
| Ref : Journal of Biological Chemistry , :101302 , 2021 |
| Abstract : Novy_2021_J.Biol.Chem__101302 |
| ESTHER : Novy_2021_J.Biol.Chem__101302 |
| PubMedSearch : Novy_2021_J.Biol.Chem__101302 |
| PubMedID: 34653507 |
| Gene_locus related to this paper: 9pezi-s4vch4 , aspfu-q4x1n0 , crysp-Q874E9 , emeni-q5b2c1 , fusso-cutas , pyrbr-Q9Y7G8 , copci-b9u443 , thite-g2rae6 , strsw-c9zcr8 |
| Title : Purification and characterization of a secretory lipolytic enzyme, MgLIP2, from Malassezia globosa - Juntachai_2011_Microbiol.(Reading)_157_3492 |
| Author(s) : Juntachai W , Oura T , Kajiwara S |
| Ref : Microbiology (Reading) , 157 :3492 , 2011 |
| Abstract : Juntachai_2011_Microbiol.(Reading)_157_3492 |
| ESTHER : Juntachai_2011_Microbiol.(Reading)_157_3492 |
| PubMedSearch : Juntachai_2011_Microbiol.(Reading)_157_3492 |
| PubMedID: 22016565 |
| Gene_locus related to this paper: malgo-a8qcw4 |
| Title : A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase - Eggert_2000_Eur.J.Biochem_267_6459 |
| Author(s) : Eggert T , Pencreac'h G , Douchet I , Verger R , Jaeger KE |
| Ref : European Journal of Biochemistry , 267 :6459 , 2000 |
| Abstract : Eggert_2000_Eur.J.Biochem_267_6459 |
| ESTHER : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedSearch : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedID: 11029590 |
| Gene_locus related to this paper: bacsu-LIPB |