P-nitrophenyl-phosphate
General
Type : Organophosphate || pNP || Chromogen
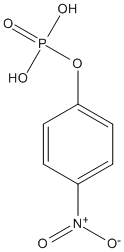
Chemical_Nomenclature : 4-nitrophenyl phosphate
Canonical SMILES : C1=CC(=CC=C1[N+](=O)[O-])OP(=O)(O)O
InChI : InChI=1S\/C6H6NO6P\/c8-7(9)5-1-3-6(4-2-5)13-14(10,11)12\/h1-4H,(H2,10,11,12)
InChIKey : XZKIHKMTEMTJQX-UHFFFAOYSA-N
Other name(s) : 4-nitrophenyl phosphate, Nitrophenylphosphate, 4-Nitrophenyl dihydrogen phosphate, P-nitrophenyl phosphate, Phosphoric acid, mono(4-nitrophenyl) ester
Target
Families : ACPH_Peptidase_S9
References (2)
| Title : Crystal Structure of an Acylpeptide Hydrolase\/Esterase from Aeropyrum pernix K1. - Bartlam_2004_Structure.(Camb)_12_1481 |
| Author(s) : Bartlam M , Wang G , Yang H , Gao R , Zhao X , Xie G , Cao S , Feng Y , Rao Z |
| Ref : Structure(Camb) , 12 :1481 , 2004 |
| Abstract : Bartlam_2004_Structure.(Camb)_12_1481 |
| ESTHER : Bartlam_2004_Structure.(Camb)_12_1481 |
| PubMedSearch : Bartlam_2004_Structure.(Camb)_12_1481 |
| PubMedID: 15296741 |
| Gene_locus related to this paper: aerpe-APE1547 |
| Title : Crystallization and preliminary crystallographic analysis of acylamino-acid releasing enzyme from the hyperthermophilic archaeon Aeropyrum pernix - Wang_2002_Acta.Crystallogr.D.Biol.Crystallogr_58_1054 |
| Author(s) : Wang G , Gao R , Ding Y , Yang H , Cao S , Feng Y , Rao Z |
| Ref : Acta Crystallographica D Biol Crystallogr , 58 :1054 , 2002 |
| Abstract : Wang_2002_Acta.Crystallogr.D.Biol.Crystallogr_58_1054 |
| ESTHER : Wang_2002_Acta.Crystallogr.D.Biol.Crystallogr_58_1054 |
| PubMedSearch : Wang_2002_Acta.Crystallogr.D.Biol.Crystallogr_58_1054 |
| PubMedID: 12037315 |
| Gene_locus related to this paper: aerpe-APE1547 |