NPC
General
Type : Carbamate || Drug || Derivative of Irinotecan || Not A\/B H target || Piperidine
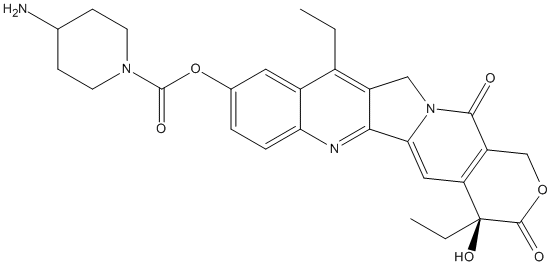
Chemical_Nomenclature : [(19S)-10,19-diethyl-19-hydroxy-14,18-dioxo-17-oxa-3,13-diazapentacyclo[11.8.0.02,11.04,9.015,20]henicosa-1(21),2,4(9),5,7,10,15(20)-heptaen-7-yl] 4-aminopiperidine-1-carboxylate
Canonical SMILES : CCC1=C2CN3C(=CC4=C(C3=O)COC(=O)C4(CC)O)C2=NC5=C1C=C(C=C5)OC(=O)N6CCC(CC6)N
InChI : InChI=1S\/C28H30N4O6\/c1-3-17-18-11-16(38-27(35)31-9-7-15(29)8-10-31)5-6-22(18)30-24-19(17)13-32-23(24)12-21-20(25(32)33)14-37-26(34)28(21,36)4-2\/h5-6,11-12,15,36H,3-4,7-10,13-14,29H2,1-2H3\/t28-\/m0\/s1
InChIKey : APWFTHDYKJHNEV-NDEPHWFRSA-N
Other name(s) : 7-ethyl-10-[4-amino-1-piperidinyl] carbonyl oxycamptothecin (NPC), 7-Ethyl-10-(4-amino-1-piperidino)carbonyloxycamptothecin, NPC (metabolite), RPR132595A, NPC, CHEBI:80552, ZINC22058728
Target
Families : Carb_B_Bacteria
References (5)
| Title : Structural insights into catalytical capability for CPT11 hydrolysis and substrate specificity of a novel marine microbial carboxylesterase, E93 - Li_2023_Front.Microbiol_13_1081094 |
| Author(s) : Li Y , Rong Z , Li Z , Cui H , Li J , Xu XW |
| Ref : Front Microbiol , 13 :1081094 , 2023 |
| Abstract : Li_2023_Front.Microbiol_13_1081094 |
| ESTHER : Li_2023_Front.Microbiol_13_1081094 |
| PubMedSearch : Li_2023_Front.Microbiol_13_1081094 |
| PubMedID: 36756200 |
| Gene_locus related to this paper: 9sphn-E93 |
| Title : Hydrolysis of irinotecan and its oxidative metabolites, 7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyloxycamptothecin and 7-ethyl-10-[4-(1-piperidino)-1-amino]-carbonyloxycamptothecin, by human carboxylesterases CES1A1, CES2, and a newly expressed carboxylesterase isoenzyme, CES3 - Sanghani_2004_Drug.Metab.Dispos_32_505 |
| Author(s) : Sanghani SP , Quinney SK , Fredenburg TB , Davis WI , Murry DJ , Bosron WF |
| Ref : Drug Metabolism & Disposition: The Biological Fate of Chemicals , 32 :505 , 2004 |
| Abstract : Sanghani_2004_Drug.Metab.Dispos_32_505 |
| ESTHER : Sanghani_2004_Drug.Metab.Dispos_32_505 |
| PubMedSearch : Sanghani_2004_Drug.Metab.Dispos_32_505 |
| PubMedID: 15100172 |
| Gene_locus related to this paper: human-CES2 , human-CES3 |
| Title : Lessons learned from the irinotecan metabolic pathway - Ma_2003_Curr.Med.Chem_10_41 |
| Author(s) : Ma MK , McLeod HL |
| Ref : Curr Med Chem , 10 :41 , 2003 |
| Abstract : Ma_2003_Curr.Med.Chem_10_41 |
| ESTHER : Ma_2003_Curr.Med.Chem_10_41 |
| PubMedSearch : Ma_2003_Curr.Med.Chem_10_41 |
| PubMedID: 12570720 |
| Title : A new metabolite of irinotecan in which formation is mediated by human hepatic cytochrome P-450 3A4 - Sai_2001_Drug.Metab.Dispos_29_1505 |
| Author(s) : Sai K , Kaniwa N , Ozawa S , Sawada JI |
| Ref : Drug Metabolism & Disposition: The Biological Fate of Chemicals , 29 :1505 , 2001 |
| Abstract : Sai_2001_Drug.Metab.Dispos_29_1505 |
| ESTHER : Sai_2001_Drug.Metab.Dispos_29_1505 |
| PubMedSearch : Sai_2001_Drug.Metab.Dispos_29_1505 |
| PubMedID: 11602529 |
| Title : Metabolism of irinotecan (CPT-11) by CYP3A4 and CYP3A5 in humans - Santos_2000_Clin.Cancer.Res_6_2012 |
| Author(s) : Santos A , Zanetta S , Cresteil T , Deroussent A , Pein F , Raymond E , Vernillet L , Risse ML , Boige V , Gouyette A , Vassal G |
| Ref : Clin Cancer Research , 6 :2012 , 2000 |
| Abstract : Santos_2000_Clin.Cancer.Res_6_2012 |
| ESTHER : Santos_2000_Clin.Cancer.Res_6_2012 |
| PubMedSearch : Santos_2000_Clin.Cancer.Res_6_2012 |
| PubMedID: 10815927 |