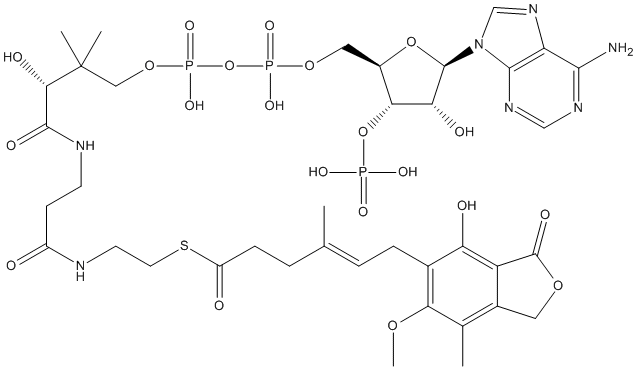
Mycophenolic-acid-CoA
General
Type : CoA || Natural || Benzofuran || Aminopurin
Chemical_Nomenclature : S-[2-[3-[[4-[[[5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] (E)-6-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1H-2-benzofuran-5-yl)-4-methylhex-4-enethioate
Canonical SMILES : CC1=C2COC(=O)C2=C(C(=C1OC)CC=C(C)CCC(=O)SCCNC(=O)CCNC(=O)C(C(C)(C)COP(=O)(O)OP(=O)(O)OCC3C(C(C(O3)N4C=NC5=C(N=CN=C54)N)O)OP(=O)(O)O)O)O
InChI : InChI=1S\/C38H54N7O21P3S\/c1-19(6-8-21-28(48)26-22(14-61-37(26)52)20(2)30(21)60-5)7-9-25(47)70-13-12-40-24(46)10-11-41-35(51)32(50)38(3,4)16-63-69(58,59)66-68(56,57)62-15-23-31(65-67(53,54)55)29(49)36(64-23)45-18-44-27-33(39)42-17-43-34(27)45\/h6,17-18,23,29,31-32,36,48-50H,7-16H2,1-5H3,(H,40,46)(H,41,51)(H,56,57)(H,58,59)(H2,39,42,43)(H2,53,54,55)\/b19-6+
InChIKey : RZGVYPYOUBIPOV-KPSZGOFPSA-N
Other name(s) : Mycophenolic acid-CoA\; (Acyl-CoA)\; [M+H]+\;, Mycophenolic acid-CoA, Mycophenolic acid-coenzyme A, Mycophenolic acid coenzyme A
MW : 1069.9
Formula : C38H54N7O21P3S
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : MpaH
References (1)
| Title : Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis - You_2021_FEBS.J_288_5768 |
| Author(s) : You C , Li F , Zhang X , Ma L , Zhang YZ , Zhang W , Li S |
| Ref : Febs J , 288 :5768 , 2021 |
| Abstract : You_2021_FEBS.J_288_5768 |
| ESTHER : You_2021_FEBS.J_288_5768 |
| PubMedSearch : You_2021_FEBS.J_288_5768 |
| PubMedID: 33843134 |
| Gene_locus related to this paper: penbr-mpaH |