Methyl-myristate
General
Type : Alkyl ester || Tetradecanoate
Chemical_Nomenclature : methyl tetradecanoate
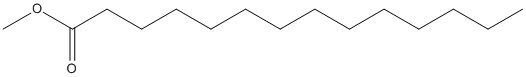
Canonical SMILES : CCCCCCCCCCCCCC(=O)OC
InChI : InChI=1S\/C15H30O2\/c1-3-4-5-6-7-8-9-10-11-12-13-14-15(16)17-2\/h3-14H2,1-2H3
InChIKey : ZAZKJZBWRNNLDS-UHFFFAOYSA-N
Other name(s) : Methyl tetradecanoate, Methyl myristate, Tetradecanoic acid, methyl ester, Uniphat A50\; Metholeneat 2495
Target
Families : No family
References (3)
| Title : The Purification and Characterization of Lipases from Lasiodiplodia theobromae, and Their Immobilization and Use for Biodiesel Production from Coconut Oil - Venkatesagowda_2018_Appl.Biochem.Biotechnol_183_619 |
| Author(s) : Venkatesagowda B , Ponugupaty E , Barbosa-Dekker AM , Dekker RFH |
| Ref : Appl Biochem Biotechnol , : , 2018 |
| Abstract : Venkatesagowda_2018_Appl.Biochem.Biotechnol_183_619 |
| ESTHER : Venkatesagowda_2018_Appl.Biochem.Biotechnol_183_619 |
| PubMedSearch : Venkatesagowda_2018_Appl.Biochem.Biotechnol_183_619 |
| PubMedID: 29250753 |
| Title : Chlamydomonas as a new organism for biodiesel production - Morowvat_2010_Bioresour.Technol_101_2059 |
| Author(s) : Morowvat MH , Rasoul-Amini S , Ghasemi Y |
| Ref : Bioresour Technol , 101 :2059 , 2010 |
| Abstract : Morowvat_2010_Bioresour.Technol_101_2059 |
| ESTHER : Morowvat_2010_Bioresour.Technol_101_2059 |
| PubMedSearch : Morowvat_2010_Bioresour.Technol_101_2059 |
| PubMedID: 19945866 |