Methyl-butyrate
General
Type : Butyrate || Alkyl ester
Chemical_Nomenclature : methyl butanoate
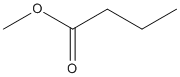
Canonical SMILES : CCCC(=O)OC
InChI : InChI=1S\/C5H10O2\/c1-3-4-5(6)7-2\/h3-4H2,1-2H3
InChIKey : UUIQMZJEGPQKFD-UHFFFAOYSA-N
Other name(s) : Methyl butanoate, Methylbutanoate, Methylbutyrate, Butanoic acid, methyl ester, Butyric acid, methyl ester, Methyl N-butyrate
Target
Families : ACHE, Carb_B_Chordata
References (4)
| Title : Characterization of LipN (Rv2970c) of Mycobacterium Tuberculosis H37Rv and its Probable Role in Xenobiotic Degradation - Jadeja_2016_J.Cell.Biochem_117_390 |
| Author(s) : Jadeja D , Dogra N , Arya S , Singh G , Kaur J |
| Ref : Journal of Cellular Biochemistry , 117 :390 , 2016 |
| Abstract : Jadeja_2016_J.Cell.Biochem_117_390 |
| ESTHER : Jadeja_2016_J.Cell.Biochem_117_390 |
| PubMedSearch : Jadeja_2016_J.Cell.Biochem_117_390 |
| PubMedID: 26212120 |
| Gene_locus related to this paper: myctu-Rv2970c |
| Title : Identification of pig liver esterase variants by tandem mass spectroscopy analysis and their characterization - Brusehaber_2007_Appl.Microbiol.Biotechnol_76_853 |
| Author(s) : Brusehaber E , Bottcher D , Musidlowska-Persson A , Albrecht D , Hecker M , Doderer K , Bornscheuer UT |
| Ref : Applied Microbiology & Biotechnology , 76 :853 , 2007 |
| Abstract : Brusehaber_2007_Appl.Microbiol.Biotechnol_76_853 |
| ESTHER : Brusehaber_2007_Appl.Microbiol.Biotechnol_76_853 |
| PubMedSearch : Brusehaber_2007_Appl.Microbiol.Biotechnol_76_853 |
| PubMedID: 17593363 |