Impranil
General
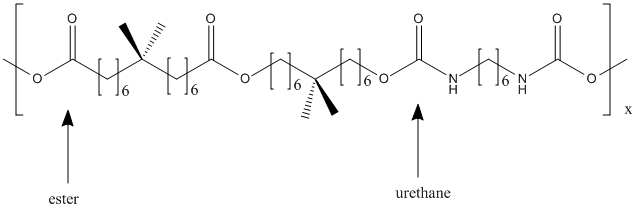
Type : Polymer || Polyurethane
Chemical_Nomenclature :
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : poly(ester)urethane, Impranil DLN(TM), Impranil DLN-SD, PEU
MW :
Formula :
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : Polyesterase-lipase-cutinase, Bacterial_lip_FamI.3, Cutinase
References (12)
| Title : Biodegradation of poly(ester-urethane) coatings by Halopseudomonas formosensis - de Witt_2024_Microb.Biotechnol_17_e14362 |
| Author(s) : de Witt J , Molitor R , Gatgens J , Ortmann de Percin Northumberland C , Kruse L , Polen T , Wynands B , van Goethem K , Thies S , Jaeger KE , Wierckx N |
| Ref : Microb Biotechnol , 17 :e14362 , 2024 |
| Abstract : de Witt_2024_Microb.Biotechnol_17_e14362 |
| ESTHER : de Witt_2024_Microb.Biotechnol_17_e14362 |
| PubMedSearch : de Witt_2024_Microb.Biotechnol_17_e14362 |
| PubMedID: 37991424 |
| Gene_locus related to this paper: 9gamm-a0a1i6bky1 |
| Title : Identification and characterization of a fungal cutinase-like enzyme CpCut1 from Cladosporium sp. P7 for polyurethane degradation - Liu_2024_Appl.Environ.Microbiol__e0147723 |
| Author(s) : Liu J , Xin K , Zhang T , Wen Y , Li D , Wei R , Zhou J , Cui Z , Dong W , Jiang M |
| Ref : Applied Environmental Microbiology , :e0147723 , 2024 |
| Abstract : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| ESTHER : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| PubMedSearch : Liu_2024_Appl.Environ.Microbiol__e0147723 |
| PubMedID: 38445906 |
| Gene_locus related to this paper: 9pezi-CpCut1 |
| Title : Biodegradation of polyurethanes by Serratia liquefaciens L135 and its polyurethanase: In silico and in vitro analyses - Salgado_2023_Environ.Pollut__122016 |
| Author(s) : Salgado CA , Silva JG , Alves de Almeida F , Dantas Vanetti MC |
| Ref : Environ Pollut , :122016 , 2023 |
| Abstract : Salgado_2023_Environ.Pollut__122016 |
| ESTHER : Salgado_2023_Environ.Pollut__122016 |
| PubMedSearch : Salgado_2023_Environ.Pollut__122016 |
| PubMedID: 37339733 |
| Gene_locus related to this paper: serli-a0a109z2v1 |
| Title : Biodegradation of polyester polyurethane by the marine fungus Cladosporium halotolerans 6UPA1 - Zhang_2022_J.Hazard.Mater_437_129406 |
| Author(s) : Zhang K , Hu J , Yang S , Xu W , Wang Z , Zhuang P , Grossart HP , Luo Z |
| Ref : J Hazard Mater , 437 :129406 , 2022 |
| Abstract : Zhang_2022_J.Hazard.Mater_437_129406 |
| ESTHER : Zhang_2022_J.Hazard.Mater_437_129406 |
| PubMedSearch : Zhang_2022_J.Hazard.Mater_437_129406 |
| PubMedID: 35753302 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : Zhang_2022_Front.Microbiol_12_803896 |
| ESTHER : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : Discovery of carbamate degrading enzymes by functional metagenomics - Ufarte_2017_PLoS.One_12_e0189201 |
| Author(s) : Ufarte L , Laville E , Duquesne S , Morgavi D , Robe P , Klopp C , Rizzo A , Pizzut-Serin S , Potocki-Veronese G |
| Ref : PLoS ONE , 12 :e0189201 , 2017 |
| Abstract : Ufarte_2017_PLoS.One_12_e0189201 |
| ESTHER : Ufarte_2017_PLoS.One_12_e0189201 |
| PubMedSearch : Ufarte_2017_PLoS.One_12_e0189201 |
| PubMedID: 29240834 |
| Gene_locus related to this paper: 9bact-CEUbrb |
| Title : Degradation of Polyester Polyurethane by Bacterial Polyester Hydrolases - Schmidt_2017_Polymers.(Basel)_9_65 |
| Author(s) : Schmidt J , Wei R , Oeser T , Dedavid e Silva LA , Breite D , Schulze A , Zimmermann W |
| Ref : Polymers (Basel) , 9 :65 , 2017 |
| Abstract : Schmidt_2017_Polymers.(Basel)_9_65 |
| ESTHER : Schmidt_2017_Polymers.(Basel)_9_65 |
| PubMedSearch : Schmidt_2017_Polymers.(Basel)_9_65 |
| PubMedID: 30970745 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 , 9bact-g9by57 , thefu-q6a0i4 , thefu-q6a0i3 |
| Title : Carbon Catabolite Repression and Impranil Polyurethane Degradation in Pseudomonas protegens Strain Pf-5 - Hung_2016_Appl.Environ.Microbiol_82_6080 |
| Author(s) : Hung CS , Zingarelli S , Nadeau LJ , Biffinger JC , Drake CA , Crouch AL , Barlow DE , Russell JN, Jr. , Crookes-Goodson WJ |
| Ref : Applied Environmental Microbiology , 82 :6080 , 2016 |
| Abstract : Hung_2016_Appl.Environ.Microbiol_82_6080 |
| ESTHER : Hung_2016_Appl.Environ.Microbiol_82_6080 |
| PubMedSearch : Hung_2016_Appl.Environ.Microbiol_82_6080 |
| PubMedID: 27496773 |
| Gene_locus related to this paper: psef5-q4kbs3 , psef5-q4kbs6 |
| Title : Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme - Howard_2012_Biodegradation_23_561 |
| Author(s) : Howard GT , Norton WN , Burks T |
| Ref : Biodegradation , 23 :561 , 2012 |
| Abstract : Howard_2012_Biodegradation_23_561 |
| ESTHER : Howard_2012_Biodegradation_23_561 |
| PubMedSearch : Howard_2012_Biodegradation_23_561 |
| PubMedID: 22228300 |
| Title : Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium - Gautam_2007_Biotechnol.Lett_29_1081 |
| Author(s) : Gautam R , Bassi AS , Yanful EK |
| Ref : Biotechnol Lett , 29 :1081 , 2007 |
| Abstract : Gautam_2007_Biotechnol.Lett_29_1081 |
| ESTHER : Gautam_2007_Biotechnol.Lett_29_1081 |
| PubMedSearch : Gautam_2007_Biotechnol.Lett_29_1081 |
| PubMedID: 17450322 |
| Title : Cloning and expression in Escherichia coli of a polyurethane-degrading enzyme from Pseudomonas fluorescens - Vega_1999_Int.Biodeterior.Biodegradation_43_49 |
| Author(s) : Vega RE , Main T , Howard GT |
| Ref : International Biodeterioration & Biodegradation , 43 :49 , 1999 |
| Abstract : Vega_1999_Int.Biodeterior.Biodegradation_43_49 |
| ESTHER : Vega_1999_Int.Biodeterior.Biodegradation_43_49 |
| PubMedSearch : Vega_1999_Int.Biodeterior.Biodegradation_43_49 |
| PubMedID: |
| Gene_locus related to this paper: psefl-PULA |
| Title : Biodegradation of a Colloidal Ester-based Polyurethane by Soil Fungi - Crabbe_1994_Int.Biodeterior.Biodegrad_33_103 |
| Author(s) : Crabbe JR , Campbell JR , Thompson L , Walz SL , Schultz WW |
| Ref : International Biodeterioration & Biodegradation , 33 :103 , 1994 |
| Abstract : Crabbe_1994_Int.Biodeterior.Biodegrad_33_103 |
| ESTHER : Crabbe_1994_Int.Biodeterior.Biodegrad_33_103 |
| PubMedSearch : Crabbe_1994_Int.Biodeterior.Biodegrad_33_103 |
| PubMedID: |