HEMT
General
Type : Terephthalate || Phthalate
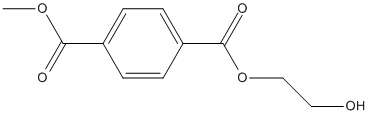
Chemical_Nomenclature : 4-O-(2-hydroxyethyl) 1-O-methyl benzene-1,4-dicarboxylate
Canonical SMILES : COC(=O)C1=CC=C(C=C1)C(=O)OCCO
InChI : InChI=1S\/C11H12O5\/c1-15-10(13)8-2-4-9(5-3-8)11(14)16-7-6-12\/h2-5,12H,6-7H2,1H3
InChIKey : DJQMYWWZWUOCBQ-UHFFFAOYSA-N
Other name(s) : 2-Hydroxyethyl methyl terephthalate, Terephthalic Acid 2-Hydroxyethyl Methyl Ester, 1,4-Benzenedicarboxylic acid, 2-hydroxyethyl methyl ester, 1,4-Benzenedicarboxylic acid, 1-(2-hydroxyethyl) 4-methyl ester, 1,4-Benzenedicarboxylicacid, 1-(2-hydroxyethyl) 4-methyl ester
Target
Families : Polyesterase-lipase-cutinase
References (6)
| Title : Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase - Fecker_2018_Biophys.J_114_1302 |
| Author(s) : Fecker T , Galaz-Davison P , Engelberger F , Narui Y , Sotomayor M , Parra LP , Ramirez-Sarmiento CA |
| Ref : Biophysical Journal , 114 :1302 , 2018 |
| Abstract : Fecker_2018_Biophys.J_114_1302 |
| ESTHER : Fecker_2018_Biophys.J_114_1302 |
| PubMedSearch : Fecker_2018_Biophys.J_114_1302 |
| PubMedID: 29590588 |
| Gene_locus related to this paper: idesa-peth |
| Title : Characterization and engineering of a plastic-degrading aromatic polyesterase - Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| Author(s) : Austin HP , Allen MD , Donohoe BS , Rorrer NA , Kearns FL , Silveira RL , Pollard BC , Dominick G , Duman R , El Omari K , Mykhaylyk V , Wagner A , Michener WE , Amore A , Skaf MS , Crowley MF , Thorne AW , Johnson CW , Woodcock HL , McGeehan JE , Beckham GT |
| Ref : Proc Natl Acad Sci U S A , 115 :E4350 , 2018 |
| Abstract : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| ESTHER : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedSearch : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedID: 29666242 |
| Gene_locus related to this paper: idesa-peth |
| Title : Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation - Joo_2018_Nat.Commun_9_382 |
| Author(s) : Joo S , Cho IJ , Seo H , Son HF , Sagong HY , Shin TJ , Choi SY , Lee SY , Kim KJ |
| Ref : Nat Commun , 9 :382 , 2018 |
| Abstract : Joo_2018_Nat.Commun_9_382 |
| ESTHER : Joo_2018_Nat.Commun_9_382 |
| PubMedSearch : Joo_2018_Nat.Commun_9_382 |
| PubMedID: 29374183 |
| Gene_locus related to this paper: idesa-peth |
| Title : New insights into the function and global distribution of polyethylene terephthalate (PET) degrading bacteria and enzymes in marine and terrestrial metagenomes - Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| Author(s) : Danso D , Schmeisser C , Chow J , Zimmermann W , Wei R , Leggewie C , Li X , Hazen T , Streit WR |
| Ref : Applied Environmental Microbiology , 84 :e2773 , 2018 |
| Abstract : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| ESTHER : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedSearch : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedID: 29427431 |
| Gene_locus related to this paper: 9burk-PET10 , 9burk-PET11 , 9gamm-a0a0d4l7e6 , 9alte-n6vy44 , 9zzzz-a0a0f9x315 , deiml-e8u721 , olean-r4ykl9 , vibga-a0a1z2siq1 , 9burk-a0a0g3bi90 , 9bact-c3ryl0 , 9actn-h6wx58 , idesa-peth , 9bact-g9by57 , acide-PBSA , morsp-lip1 |
| Title : Structural insight into catalytic mechanism of PET hydrolase - Han_2017_Nat.Commun_8_2106 |
| Author(s) : Han X , Liu W , Huang JW , Ma J , Zheng Y , Ko TP , Xu L , Cheng YS , Chen CC , Guo RT |
| Ref : Nat Commun , 8 :2106 , 2017 |
| Abstract : Han_2017_Nat.Commun_8_2106 |
| ESTHER : Han_2017_Nat.Commun_8_2106 |
| PubMedSearch : Han_2017_Nat.Commun_8_2106 |
| PubMedID: 29235460 |
| Gene_locus related to this paper: idesa-peth |
| Title : Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76A resolution - Kitadokoro_2012_Polym.Degrad.Stab_97_771 |
| Author(s) : Kitadokoro K , Thumarat U , Nakamura R , Nishimura K , Karatani H , Suzuki H , Kawai F |
| Ref : Polymer Degradation and Stability , 97 :771 , 2012 |
| Abstract : Kitadokoro_2012_Polym.Degrad.Stab_97_771 |
| ESTHER : Kitadokoro_2012_Polym.Degrad.Stab_97_771 |
| PubMedSearch : Kitadokoro_2012_Polym.Degrad.Stab_97_771 |
| PubMedID: |
| Gene_locus related to this paper: 9acto-f7ix06 |