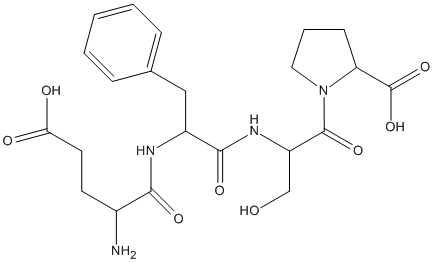
Glu-Phe-Ser-Pro
General
Type : Peptide || Pyrrolidine
Chemical_Nomenclature : 1-[2-[[2-[(2-amino-4-carboxybutanoyl)amino]-3-phenylpropanoyl]amino]-3-hydroxypropanoyl]pyrrolidine-2-carboxylic acid
Canonical SMILES : C1CC(N(C1)C(=O)C(CO)NC(=O)C(CC2=CC=CC=C2)NC(=O)C(CCC(=O)O)N)C(=O)O
InChI : InChI=1S\/C22H30N4O8\/c23-14(8-9-18(28)29)19(30)24-15(11-13-5-2-1-3-6-13)20(31)25-16(12-27)21(32)26-10-4-7-17(26)22(33)34\/h1-3,5-6,14-17,27H,4,7-12,23H2,(H,24,30)(H,25,31)(H,28,29)(H,33,34)
InChIKey : ZHGNQCAJUQNOTB-UHFFFAOYSA-N
Other name(s) : EFSP
MW : 478.50
Formula : C22H30N4O8
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : S9N_PPCE_Peptidase_S9
References (1)
| Title : Electrostatic environment at the active site of prolyl oligopeptidase is highly influential during substrate binding - Szeltner_2003_J.Biol.Chem_278_48786 |
| Author(s) : Szeltner Z , Rea D , Renner V , Juliano L , Fulop V , Polgar L |
| Ref : Journal of Biological Chemistry , 278 :48786 , 2003 |
| Abstract : Szeltner_2003_J.Biol.Chem_278_48786 |
| ESTHER : Szeltner_2003_J.Biol.Chem_278_48786 |
| PubMedSearch : Szeltner_2003_J.Biol.Chem_278_48786 |
| PubMedID: 14514675 |
| Gene_locus related to this paper: pig-ppce |