Ethylbenzoate
General
Type : Aryl ester || Benzoate
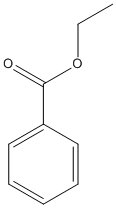
Chemical_Nomenclature : ethyl benzoate
Canonical SMILES : CCOC(=O)C1=CC=CC=C1
InChI : InChI=1S\/C9H10O2\/c1-2-11-9(10)8-6-4-3-5-7-8\/h3-7H,2H2,1H3
InChIKey : MTZQAGJQAFMTAQ-UHFFFAOYSA-N
Other name(s) : Ethyl benzoate, Ethyl-benzoate, Benzoic acid, ethyl ester, Benzoic ether, Ethyl benzenecarboxylate
Target
Families : Canar_LipB, 6_AlphaBeta_hydrolase
References (3)
| Title : An enzyme from Auricularia auricula-judae combining both benzoyl and cinnamoyl esterase activity - Haase-Aschoff_2013_Process.Biochem_48_1872 |
| Author(s) : Haase-Aschoff P , Linke D , Nimtz M , Popper L , Berger RG |
| Ref : Process Biochemistry , 48 :1872 , 2013 |
| Abstract : Haase-Aschoff_2013_Process.Biochem_48_1872 |
| ESTHER : Haase-Aschoff_2013_Process.Biochem_48_1872 |
| PubMedSearch : Haase-Aschoff_2013_Process.Biochem_48_1872 |
| PubMedID: |
| Gene_locus related to this paper: aurst-EJD51015 |
| Title : Understanding the plasticity of the alpha\/beta hydrolase fold: lid swapping on the Candida antarctica lipase B results in chimeras with interesting biocatalytic properties - Skjot_2009_Chembiochem_10_520 |
| Author(s) : Skjot M , De Maria L , Chatterjee R , Svendsen A , Patkar SA , Ostergaard PR , Brask J |
| Ref : Chembiochem , 10 :520 , 2009 |
| Abstract : Skjot_2009_Chembiochem_10_520 |
| ESTHER : Skjot_2009_Chembiochem_10_520 |
| PubMedSearch : Skjot_2009_Chembiochem_10_520 |
| PubMedID: 19156649 |
| Gene_locus related to this paper: canar-LipB |