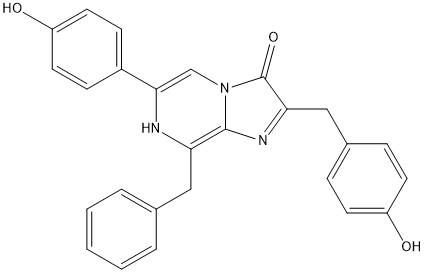
Coelenterazine
General
Type : Luciferin || Natural
Chemical_Nomenclature : N-[2-benzyl-6-(4-oxocyclohexa-2,5-dien-1-ylidene)-1H-pyrazin-3-yl]-2-(4-hydroxyphenyl)acetamide
Canonical SMILES : C1=CC=C(C=C1)CC2=C(N=CC(=C3C=CC(=O)C=C3)N2)NC(=O)CC4=CC=C(C=C4)O
InChI : InChI=1S\/C25H21N3O3\/c29-20-10-6-18(7-11-20)15-24(31)28-25-22(14-17-4-2-1-3-5-17)27-23(16-26-25)19-8-12-21(30)13-9-19\/h1-13,16,27,29H,14-15H2,(H,28,31)
InChIKey : YHIPILPTUVMWQT-UHFFFAOYSA-N
Other name(s) : Oplophorus luciferin, 55779-48-1, CLZN, CTZ, Coelenterazine ,native
MW : 423.46
Formula : C26H21N3O3
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : Haloalkane_dehalogenase-HLD2
References (4)
| Title : Deciphering Enzyme Mechanisms with Engineered Ancestors and Substrate Analogues - Gao_2023_ChemCatChem_15_e202300745 |
| Author(s) : Gao T , Damborsky J , Janin YL , Marek M |
| Ref : ChemCatChem , 15 :e202300745 , 2023 |
| Abstract : Gao_2023_ChemCatChem_15_e202300745 |
| ESTHER : Gao_2023_ChemCatChem_15_e202300745 |
| PubMedSearch : Gao_2023_ChemCatChem_15_e202300745 |
| PubMedID: |
| Gene_locus related to this paper: renre-luc |
| Title : Light-Emitting Dehalogenases: Reconstruction of Multifunctional Biocatalysts - Chaloupkova_2019_ACS.Catal_9_48103 |
| Author(s) : Chaloupkova, R , Liskova V , Tool M , Markova K , Sebestova E , Hernychova L , Marek M , Pinto GP , Pluskal D , Waterman J , Prokop Z , Damborsky J |
| Ref : ACS Catal , 9 :4810 , 2019 |
| Abstract : Chaloupkova_2019_ACS.Catal_9_48103 |
| ESTHER : Chaloupkova_2019_ACS.Catal_9_48103 |
| PubMedSearch : Chaloupkova_2019_ACS.Catal_9_48103 |
| PubMedID: |
| Gene_locus related to this paper: 9zzzz-AncHLDRLuc2 , 9zzzz-AncHLDRLuc , renre-luc |
| Title : Structure-function studies on the active site of the coelenterazine-dependent luciferase from Renilla - Woo_2008_Protein.Sci_17_725 |
| Author(s) : Woo J , Howell MH , von Arnim AG |
| Ref : Protein Science , 17 :725 , 2008 |
| Abstract : Woo_2008_Protein.Sci_17_725 |
| ESTHER : Woo_2008_Protein.Sci_17_725 |
| PubMedSearch : Woo_2008_Protein.Sci_17_725 |
| PubMedID: 18359861 |
| Gene_locus related to this paper: renre-luc |
| Title : Crystal structures of the luciferase and green fluorescent protein from Renilla reniformis - Loening_2007_J.Mol.Biol_374_1017 |
| Author(s) : Loening AM , Fenn TD , Gambhir SS |
| Ref : Journal of Molecular Biology , 374 :1017 , 2007 |
| Abstract : Loening_2007_J.Mol.Biol_374_1017 |
| ESTHER : Loening_2007_J.Mol.Biol_374_1017 |
| PubMedSearch : Loening_2007_J.Mol.Biol_374_1017 |
| PubMedID: 17980388 |
| Gene_locus related to this paper: renre-luc |