4,9-DHSA
General
Type : Inden
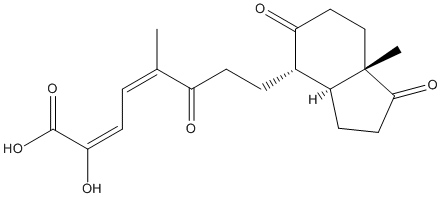
Chemical_Nomenclature : (2E,4Z)-8-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-2,3,3a,4,6,7-hexahydroinden-4-yl]-1-hydroxy-5-methyl-1,6-dioxoocta-2,4-dien-2-olate, (2E,4Z)-8-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-2,3,3a,4,6,7-hexahydroinden-4-yl]-2-hydroxy-5-methyl-6-oxoocta-2,4-dienoic acid
Canonical SMILES : CC(=CC=C(C(=O)O)[O-])C(=O)CCC1C2CCC(=O)C2(CCC1=O)C || CC(=CC=C(C(=O)O)O)C(=O)CCC1C2CCC(=O)C2(CCC1=O)C
InChI : InChI=1S\/C19H24O6\/c1-11(3-6-16(22)18(24)25)14(20)7-4-12-13-5-8-17(23)19(13,2)10-9-15(12)21\/h3,6,12-13,22H,4-5,7-10H2,1-2H3,(H,24,25)\/p-1\/b11-3-,16-6+\/t12-,13-,19-\/m0\/s1 || InChI=1S\/C19H24O6\/c1-11(3-6-16(22)18(24)25)14(20)7-4-12-13-5-8-17(23)19(13,2)10-9-15(12)21\/h3,6,12-13,22H,4-5,7-10H2,1-2H3,(H,24,25)\/b11-3-,16-6+\/t12-,13-,19-\/m0\/s1
InChIKey : HRJXKKSJPNWKCP-PBEJIIKFSA-M || HRJXKKSJPNWKCP-PBEJIIKFSA-N
Other name(s) : CHEBI:58607, 5-dioxooctahydro-1H-inden-4-yl]-6-oxoocta-2,4-dienoate, (3E,1Z)-4,5-9,10-Diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate, 3-hydroxy-5,9,17-trioxo-4,5:9,10-disecoandrosta-1(10),2-dien-4-oic acid, 3-Hydroxy-5,9,17-trioxo-4,5:9,10-disecoandrosta-1(10),2-dien-4-oate, CHEBI:29086, (2E,4Z)-8-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-2,3,3a,4,6,7-hexahydroinden-4-yl]-2-hydroxy-5-methyl-6-oxoocta-2,4-dienoic acid
MW : 347.4 || 348.4
Formula : C19H23O6- || C19H24O6
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : Carbon-carbon_bond_hydrolase
References (5)
| Title : Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism - Lack_2010_J.Biol.Chem_285_434 |
| Author(s) : Lack NA , Yam KC , Lowe ED , Horsman GP , Owen RL , Sim E , Eltis LD |
| Ref : Journal of Biological Chemistry , 285 :434 , 2010 |
| Abstract : Lack_2010_J.Biol.Chem_285_434 |
| ESTHER : Lack_2010_J.Biol.Chem_285_434 |
| PubMedSearch : Lack_2010_J.Biol.Chem_285_434 |
| PubMedID: 19875455 |
| Gene_locus related to this paper: myctu-Rv3569c |
| Title : Structure of HsaD, a steroid-degrading hydrolase, from Mycobacterium tuberculosis - Lack_2008_Acta.Crystallogr.Sect.F.Struct.Biol.Cryst.Commun_64_2 |
| Author(s) : Lack N , Lowe ED , Liu J , Eltis LD , Noble ME , Sim E , Westwood IM |
| Ref : Acta Crystallographica Sect F Struct Biol Cryst Commun , 64 :2 , 2008 |
| Abstract : Lack_2008_Acta.Crystallogr.Sect.F.Struct.Biol.Cryst.Commun_64_2 |
| ESTHER : Lack_2008_Acta.Crystallogr.Sect.F.Struct.Biol.Cryst.Commun_64_2 |
| PubMedSearch : Lack_2008_Acta.Crystallogr.Sect.F.Struct.Biol.Cryst.Commun_64_2 |
| PubMedID: 18097091 |
| Gene_locus related to this paper: myctu-Rv3569c |
| Title : A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages - Van der Geize_2007_Proc.Natl.Acad.Sci.U.S.A_104_1947 |
| Author(s) : Van der Geize R , Yam K , Heuser T , Wilbrink MH , Hara H , Anderton MC , Sim E , Dijkhuizen L , Davies JE , Mohn WW , Eltis LD |
| Ref : Proc Natl Acad Sci U S A , 104 :1947 , 2007 |
| Abstract : Van der Geize_2007_Proc.Natl.Acad.Sci.U.S.A_104_1947 |
| ESTHER : Van der Geize_2007_Proc.Natl.Acad.Sci.U.S.A_104_1947 |
| PubMedSearch : Van der Geize_2007_Proc.Natl.Acad.Sci.U.S.A_104_1947 |
| PubMedID: 17264217 |
| Gene_locus related to this paper: myctu-Rv3569c |
| Title : Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441 - Horinouchi_2005_Appl.Environ.Microbiol_71_5275 |
| Author(s) : Horinouchi M , Hayashi T , Koshino H , Kurita T , Kudo T |
| Ref : Applied Environmental Microbiology , 71 :5275 , 2005 |
| Abstract : Horinouchi_2005_Appl.Environ.Microbiol_71_5275 |
| ESTHER : Horinouchi_2005_Appl.Environ.Microbiol_71_5275 |
| PubMedSearch : Horinouchi_2005_Appl.Environ.Microbiol_71_5275 |
| PubMedID: 16151114 |
| Gene_locus related to this paper: comte-TESD |
| Title : Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR - Horinouchi_2004_Biochem.Biophys.Res.Commun_324_597 |
| Author(s) : Horinouchi M , Kurita T , Yamamoto T , Hatori E , Hayashi T , Kudo T |
| Ref : Biochemical & Biophysical Research Communications , 324 :597 , 2004 |
| Abstract : Horinouchi_2004_Biochem.Biophys.Res.Commun_324_597 |
| ESTHER : Horinouchi_2004_Biochem.Biophys.Res.Commun_324_597 |
| PubMedSearch : Horinouchi_2004_Biochem.Biophys.Res.Commun_324_597 |
| PubMedID: 15474469 |