3-Methyl-3-butenyl-acetate
General
Type : Natural || Acetate
Chemical_Nomenclature : 3-methylbut-3-enyl acetate
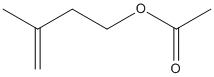
Canonical SMILES : CC(=C)CCOC(=O)C
InChI : InChI=1S\/C7H12O2\/c1-6(2)4-5-9-7(3)8\/h1,4-5H2,2-3H3
InChIKey : OCUAPVNNQFAQSM-UHFFFAOYSA-N
Other name(s) : 3-Methyl-3-buten-1-yl acetate, 3-Methyl-3-butenyl acetate, 3-methylbut-3-enyl acetate, 3-Buten-1-ol, 3-methyl-, acetate, isopentenyl acetate, 3-Buten-1-ol, 3-methyl-, 1-acetate, 3-Methyl-3-buten-1-ol, acetate
Target
Families : No family
References (3)
| Title : Female-biased attraction of Oriental fruit fly, bactrocera dorsalis (Hendel), to a blend of host fruit volatiles from Terminalia catappa L - Siderhurst_2006_J.Chem.Ecol_32_2513 |
| Author(s) : Siderhurst MS , Jang EB |
| Ref : J Chem Ecol , 32 :2513 , 2006 |
| Abstract : Siderhurst_2006_J.Chem.Ecol_32_2513 |
| ESTHER : Siderhurst_2006_J.Chem.Ecol_32_2513 |
| PubMedSearch : Siderhurst_2006_J.Chem.Ecol_32_2513 |
| PubMedID: 17082987 |
| Title : Chemistry of fruit flies: Glandular secretion ofBactrocera (Polistomimetes) visenda (Hardy) - Krohn_1992_J.Chem.Ecol_18_2169 |
| Author(s) : Krohn S , Fletcher MT , Kitching W , Moore CJ , Drew RA , Francke W |
| Ref : J Chem Ecol , 18 :2169 , 1992 |
| Abstract : Krohn_1992_J.Chem.Ecol_18_2169 |
| ESTHER : Krohn_1992_J.Chem.Ecol_18_2169 |
| PubMedSearch : Krohn_1992_J.Chem.Ecol_18_2169 |
| PubMedID: 24254865 |