PHB
General
Type : Natural || Polymer || Polyhydroxyalkanoate
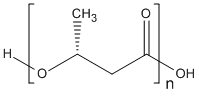
Chemical_Nomenclature : poly(3-hydroxybutyrate)
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : P(3HB), P3HB, poly(3-hydroxybutyrate), polyhydroxybutyrate, acide 3-hydroxybutyrique homopolymer, poly(acide-3-hydroxybutyrique), C06143
References (10)
| Title : Purification, characterization, and gene cloning of an Aspergillus fumigatus polyhydroxybutyrate depolymerase used for degradation of polyhydroxybutyrate, polyethylene succinate, and polybutylene succinate - Jung_2018_Polym.Degrad.Stab_154_186 |
| Author(s) : Jung HW , Yang MK , Su RC |
| Ref : Polymer Degradation and Stability , 154 :154 , 2018 |
| Abstract : Jung_2018_Polym.Degrad.Stab_154_186 |
| ESTHER : Jung_2018_Polym.Degrad.Stab_154_186 |
| PubMedSearch : Jung_2018_Polym.Degrad.Stab_154_186 |
| PubMedID: |
| Gene_locus related to this paper: aspfu-q4w9v8 |
| Title : Biochemical analysis and structure determination of Paucimonas lemoignei poly(3-hydroxybutyrate) (PHB) depolymerase PhaZ7 muteins reveal the PHB binding site and details of substrate-enzyme interactions - Jendrossek_2013_Mol.Microbiol_90_649 |
| Author(s) : Jendrossek D , Hermawan S , Subedi B , Papageorgiou AC |
| Ref : Molecular Microbiology , 90 :649 , 2013 |
| Abstract : Jendrossek_2013_Mol.Microbiol_90_649 |
| ESTHER : Jendrossek_2013_Mol.Microbiol_90_649 |
| PubMedSearch : Jendrossek_2013_Mol.Microbiol_90_649 |
| PubMedID: 24007310 |
| Gene_locus related to this paper: psele-PHAZ7 |
| Title : Evaluation of immobilized lipases on poly-hydroxybutyrate beads to catalyze biodiesel synthesis - Mendes_2012_Int.J.Biol.Macromol_50_503 |
| Author(s) : Mendes AA , Oliveira PC , Velez AM , Giordano RC , Giordano Rde L , de Castro HF |
| Ref : Int J Biol Macromol , 50 :503 , 2012 |
| Abstract : Mendes_2012_Int.J.Biol.Macromol_50_503 |
| ESTHER : Mendes_2012_Int.J.Biol.Macromol_50_503 |
| PubMedSearch : Mendes_2012_Int.J.Biol.Macromol_50_503 |
| PubMedID: 22285987 |
| Title : The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters - Hisano_2006_J.Mol.Biol_356_993 |
| Author(s) : Hisano T , Kasuya K , Tezuka Y , Ishii N , Kobayashi T , Shiraki M , Oroudjev E , Hansma H , Iwata T , Doi Y , Saito T , Miki K |
| Ref : Journal of Molecular Biology , 356 :993 , 2006 |
| Abstract : Hisano_2006_J.Mol.Biol_356_993 |
| ESTHER : Hisano_2006_J.Mol.Biol_356_993 |
| PubMedSearch : Hisano_2006_J.Mol.Biol_356_993 |
| PubMedID: 16405909 |
| Gene_locus related to this paper: penfu-PHAZ |
| Title : A novel PHB depolymerase from a thermophilic Streptomyces sp - Calabia_2006_Biotechnol.Lett_28_383 |
| Author(s) : Calabia BP , Tokiwa Y |
| Ref : Biotechnol Lett , 28 :383 , 2006 |
| Abstract : Calabia_2006_Biotechnol.Lett_28_383 |
| ESTHER : Calabia_2006_Biotechnol.Lett_28_383 |
| PubMedSearch : Calabia_2006_Biotechnol.Lett_28_383 |
| PubMedID: 16614903 |
| Title : Repeated batch cultivation of Ralstonia eutropha for Poly (beta-hydroxybutyrate) production - Khanna_2005_Biotechnol.Lett_27_1401 |
| Author(s) : Khanna S , Srivastava AK |
| Ref : Biotechnol Lett , 27 :1401 , 2005 |
| Abstract : Khanna_2005_Biotechnol.Lett_27_1401 |
| ESTHER : Khanna_2005_Biotechnol.Lett_27_1401 |
| PubMedSearch : Khanna_2005_Biotechnol.Lett_27_1401 |
| PubMedID: 16215857 |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| ESTHER : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Ralstonia eutropha H16 encodes two and possibly three intracellular Poly[D-(-)-3-hydroxybutyrate] depolymerase genes - York_2003_J.Bacteriol_185_3788 |
| Author(s) : York GM , Lupberger J , Tian J , Lawrence AG , Stubbe J , Sinskey AJ |
| Ref : Journal of Bacteriology , 185 :3788 , 2003 |
| Abstract : York_2003_J.Bacteriol_185_3788 |
| ESTHER : York_2003_J.Bacteriol_185_3788 |
| PubMedSearch : York_2003_J.Bacteriol_185_3788 |
| PubMedID: 12813072 |
| Gene_locus related to this paper: cupne-q7wt48 , cupne-q7wt49 |
| Title : A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids - Handrick_2001_J.Biol.Chem_276_36215 |
| Author(s) : Handrick R , Reinhardt S , Focarete ML , Scandola M , Adamus G , Kowalczuk M , Jendrossek D |
| Ref : Journal of Biological Chemistry , 276 :36215 , 2001 |
| Abstract : Handrick_2001_J.Biol.Chem_276_36215 |
| ESTHER : Handrick_2001_J.Biol.Chem_276_36215 |
| PubMedSearch : Handrick_2001_J.Biol.Chem_276_36215 |
| PubMedID: 11457823 |
| Gene_locus related to this paper: psele-PHAZ7 |
| Title : Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate-binding domains - Ohura_1999_Appl.Environ.Microbiol_65_189 |
| Author(s) : Ohura T , Kasuya KI , Doi Y |
| Ref : Applied Environmental Microbiology , 65 :189 , 1999 |
| Abstract : Ohura_1999_Appl.Environ.Microbiol_65_189 |
| ESTHER : Ohura_1999_Appl.Environ.Microbiol_65_189 |
| PubMedSearch : Ohura_1999_Appl.Environ.Microbiol_65_189 |
| PubMedID: 9872779 |
| Gene_locus related to this paper: psesp-PHBDP1 |