tetramethylrhodamine-HaloTag-ligand
General
Type : Fluorescent Probe,HaloTag-ligand,Rhodamine derivative,Xanthene
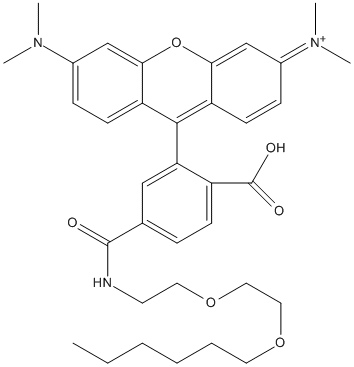
Chemical_Nomenclature : [9-[2-carboxy-5-[2-(2-hexoxyethoxy)ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethylazanium
Canonical SMILES : CCCCCCOCCOCCNC(=O)C1=CC(=C(C=C1)C(=O)O)C2=C3C=CC(=[N+](C)C)C=C3OC4=C2C=CC(=C4)N(C)C
InChI : InChI=1S\/C35H43N3O6\/c1-6-7-8-9-17-42-19-20-43-18-16-36-34(39)24-10-13-27(35(40)41)30(21-24)33-28-14-11-25(37(2)3)22-31(28)44-32-23-26(38(4)5)12-15-29(32)33\/h10-15,21-23H,6-9,16-20H2,1-5H3,(H-,36,39,40,41)\/p+1
InChIKey : HKVFTIWQXRNXNR-UHFFFAOYSA-O
Other name(s) : PVY
MW : 602.7
Formula : C35H44N3O6+
CAS_number :
PubChem : 154700563
UniChem : HKVFTIWQXRNXNR-UHFFFAOYSA-O
IUPHAR :
Wikipedia :
Target
Families : tetramethylrhodamine-HaloTag-ligand ligand of proteins in family: Haloalkane_dehalogenase-HLD2
Stucture : 6U32 Crystal structure of HaloTag bound to tetramethylrhodamine-HaloTag ligand
Protein : rhoso-halo1
References (8)
| Title : The HaloTag as a general scaffold for far-red tunable chemigenetic indicators - Deo_2021_Nat.Chem.Biol_17_718 |
| Author(s) : Deo C , Abdelfattah AS , Bhargava HK , Berro AJ , Falco N , Farrants H , Moeyaert B , Chupanova M , Lavis LD , Schreiter ER |
| Ref : Nat Chemical Biology , 17 :718 , 2021 |
| Abstract : Deo_2021_Nat.Chem.Biol_17_718 |
| ESTHER : Deo_2021_Nat.Chem.Biol_17_718 |
| PubMedSearch : Deo_2021_Nat.Chem.Biol_17_718 |
| PubMedID: 33795886 |
| Gene_locus related to this paper: rhoso-halo1 |
| Title : A robust and economical pulse-chase protocol to measure the turnover of HaloTag fusion proteins - Merrill_2019_J.Biol.Chem_294_16164 |
| Author(s) : Merrill RA , Song J , Kephart RA , Klomp AJ , Noack CE , Strack S |
| Ref : Journal of Biological Chemistry , 294 :16164 , 2019 |
| Abstract : Merrill_2019_J.Biol.Chem_294_16164 |
| ESTHER : Merrill_2019_J.Biol.Chem_294_16164 |
| PubMedSearch : Merrill_2019_J.Biol.Chem_294_16164 |
| PubMedID: 31511325 |
| Title : Quantification of very low-abundant proteins in bacteria using the HaloTag and epi-fluorescence microscopy - Lepore_2019_Sci.Rep_9_7902 |
| Author(s) : Lepore A , Taylor H , Landgraf D , Okumus B , Jaramillo-Riveri S , McLaren L , Bakshi S , Paulsson J , Karoui ME |
| Ref : Sci Rep , 9 :7902 , 2019 |
| Abstract : Lepore_2019_Sci.Rep_9_7902 |
| ESTHER : Lepore_2019_Sci.Rep_9_7902 |
| PubMedSearch : Lepore_2019_Sci.Rep_9_7902 |
| PubMedID: 31133640 |
| Title : Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy - Liss_2015_Sci.Rep_5_17740 |
| Author(s) : Liss V , Barlag B , Nietschke M , Hensel M |
| Ref : Sci Rep , 5 :17740 , 2015 |
| Abstract : Liss_2015_Sci.Rep_5_17740 |
| ESTHER : Liss_2015_Sci.Rep_5_17740 |
| PubMedSearch : Liss_2015_Sci.Rep_5_17740 |
| PubMedID: 26643905 |
| Title : Photoswitching-free FRAP analysis with a genetically encoded fluorescent tag - Morisaki_2014_PLoS.One_9_e107730 |
| Author(s) : Morisaki T , McNally JG |
| Ref : PLoS ONE , 9 :e107730 , 2014 |
| Abstract : Morisaki_2014_PLoS.One_9_e107730 |
| ESTHER : Morisaki_2014_PLoS.One_9_e107730 |
| PubMedSearch : Morisaki_2014_PLoS.One_9_e107730 |
| PubMedID: 25233348 |
| Title : Quantitative in vivo fluorescence cross-correlation analyses highlight the importance of competitive effects in the regulation of protein-protein interactions - Sadaie_2014_Mol.Cell.Biol_34_3272 |
| Author(s) : Sadaie W , Harada Y , Matsuda M , Aoki K |
| Ref : Molecular & Cellular Biology , 34 :3272 , 2014 |
| Abstract : Sadaie_2014_Mol.Cell.Biol_34_3272 |
| ESTHER : Sadaie_2014_Mol.Cell.Biol_34_3272 |
| PubMedSearch : Sadaie_2014_Mol.Cell.Biol_34_3272 |
| PubMedID: 24958104 |
| Title : Optically detected structural change in the N-terminal region of the voltage-sensor domain - Tsutsui_2013_Biophys.J_105_108 |
| Author(s) : Tsutsui H , Jinno Y , Tomita A , Okamura Y |
| Ref : Biophysical Journal , 105 :108 , 2013 |
| Abstract : Tsutsui_2013_Biophys.J_105_108 |
| ESTHER : Tsutsui_2013_Biophys.J_105_108 |
| PubMedSearch : Tsutsui_2013_Biophys.J_105_108 |
| PubMedID: 23823229 |
| Title : A HaloTag method for assessing the retrograde axonal transport of the p75 neurotrophin receptor and other proteins in compartmented cultures of rat sympathetic neurons - Mok_2013_J.Neurosci.Methods_214_91 |
| Author(s) : Mok SA , Lund K , Lapointe P , Campenot RB |
| Ref : Journal of Neuroscience Methodsods , 214 :91 , 2013 |
| Abstract : Mok_2013_J.Neurosci.Methods_214_91 |
| ESTHER : Mok_2013_J.Neurosci.Methods_214_91 |
| PubMedSearch : Mok_2013_J.Neurosci.Methods_214_91 |
| PubMedID: 23348044 |