W22
A precursor of Lotrafiban ( a potent non-peptidic antagonist that inhibits platelet aggregation). The S enantiomer is necessary for activity.Hydrolysis of racemic W22-Acetate by Lipase Novozyme 435 results in only the S isomer product
General
Type : Benzodiazepin,Diazepan,Azepane
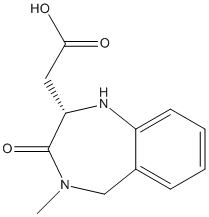
Chemical_Nomenclature : 2-[(2S)-4-methyl-3-oxo-2,5-dihydro-1H-1,4-benzodiazepin-2-yl]acetic acid
Canonical SMILES : CN1CC2=CC=CC=C2N[C@H](C1=O)CC(=O)O
InChI : InChI=1S\/C12H14N2O3\/c1-14-7-8-4-2-3-5-9(8)13-10(12(14)17)6-11(15)16\/h2-5,10,13H,6-7H2,1H3,(H,15,16)\/t10-\/m0\/s1
InChIKey : CLWDLBDPVUWYEW-JTQLQIEISA-N
Other name(s) : [(2S)-4-methyl-3-oxo-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-2-yl]acetic acid,SCHEMBL8503077,DB08717,(2S)-3(2H)-Oxo-4-methyl-4,5-dihydro-1H-1,4-benzodiazepine-2beta-acetic acid
MW : 234.25
Formula : C12H14N2O3
CAS_number :
PubChem : 11117974
UniChem : CLWDLBDPVUWYEW-JTQLQIEISA-N
IUPHAR :
Wikipedia :
Target
Families : W22 ligand of proteins in family: Bacterial_esterase
Stucture : 2WKW Alcaligenes esterase complexed with product analogue
Protein : alcsp-cxest
References (2)
| Title : Lipase catalysed resolution of the Lotrafiban intermediate 2,3,4,5-tetrahydro-4-methyl-3-oxo-1 H-1,4-benzodiazepine-2-acetic acid methyl ester in ionic liquids: comparison to the industrial t-butanol process - Roberts_2004_Green.Chem_6_475 |
| Author(s) : Roberts NJ , Seago A , Carey JS , Freer R , Preston C , Lye GJ |
| Ref : Green Chem , 6 :475 , 2004 |
| Abstract : Roberts_2004_Green.Chem_6_475 |
| ESTHER : Roberts_2004_Green.Chem_6_475 |
| PubMedSearch : Roberts_2004_Green.Chem_6_475 |
| PubMedID: |
| Gene_locus related to this paper: canar-LipB |
| Title : The atomic-resolution structure of a novel bacterial esterase - Bourne_2000_Structure.Fold.Des_8_143 |
| Author(s) : Bourne PC , Isupov MN , Littlechild JA |
| Ref : Structure Fold Des , 8 :143 , 2000 |
| Abstract : Bourne_2000_Structure.Fold.Des_8_143 |
| ESTHER : Bourne_2000_Structure.Fold.Des_8_143 |
| PubMedSearch : Bourne_2000_Structure.Fold.Des_8_143 |
| PubMedID: 10673440 |
| Gene_locus related to this paper: alcsp-cxest |