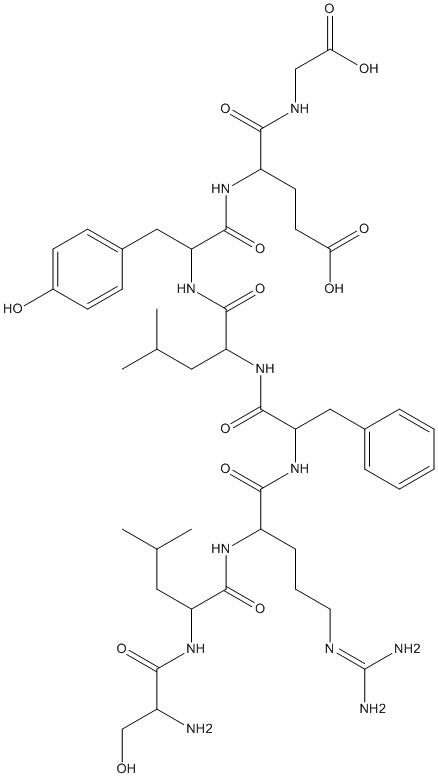
SLRFLYEG
SUMO1 was identified as an interacting partner of DPP8 and DPP9. The region of interaction on SUMO1 is the E67 interacting loop (EIL). EIL peptide was able to compete with the interaction of SUMO1 with DPP8 or DPP9. EIL also exhibited an inhibitory effect on DPP8/9. Systematic variation in the EIL amino acid sequence gave a strong inhibitor, SLRFLYEG
General
Type : Peptide
Chemical_Nomenclature : (4S)-4-[[(2S)-2-[[(2S)-2-[[(2S)-2-[[(2S)-2-[[(2S)-2-[[(2S)-2-amino-3-hydroxypropanoyl]amino]-4-methylpentanoyl]amino]-5-(diaminomethylideneamino)pentanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]-3-(4-hydroxyphenyl)propanoyl]amino]-5-(carboxymethylamino)-5-oxopentanoic acid
Canonical SMILES : CC(C)CC(C(=O)NC(CC1=CC=C(C=C1)O)C(=O)NC(CCC(=O)O)C(=O)NCC(=O)O)NC(=O)C(CC2=CC=CC=C2)NC(=O)C(CCCN=C(N)N)NC(=O)C(CC(C)C)NC(=O)C(CO)N
InChI : InChI=1S\/C46H69N11O13\/c1-25(2)19-33(54-39(64)30(47)24-58)42(67)52-31(11-8-18-50-46(48)49)41(66)56-35(21-27-9-6-5-7-10-27)45(70)55-34(20-26(3)4)43(68)57-36(22-28-12-14-29(59)15-13-28)44(69)53-32(16-17-37(60)61)40(65)51-23-38(62)63\/h5-7,9-10,12-15,25-26,30-36,58-59H,8,11,16-24,47H2,1-4H3,(H,51,65)(H,52,67)(H,53,69)(H,54,64)(H,55,70)(H,56,66)(H,57,68)(H,60,61)(H,62,63)(H4,48,49,50)\/t30-,31-,32-,33-,34-,35-,36-\/m0\/s1
InChIKey : JWANCNBNXTWMNU-QJCLFNHPSA-N
Other name(s) : SER-LEU-ARG-PHE-LEU-TYR-GLU-GLY,Slrflyeg,BDBM213831,ZINC208939526
MW : 984.12
Formula : C46H69N11O13
CAS_number :
PubChem : 73774647
UniChem : JWANCNBNXTWMNU-QJCLFNHPSA-N
IUPHAR :
Wikipedia :
Target
Families : SLRFLYEG ligand of proteins in family: DPP4N_Peptidase_S9
Stucture :
Protein : human-DPP9
References (3)
| Title : Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer - Ross_2018_Proc.Natl.Acad.Sci.U.S.A_115_E1437 |
| Author(s) : Ross B , Krapp S , Augustin M , Kierfersauer R , Arciniega M , Geiss-Friedlander R , Huber R |
| Ref : Proc Natl Acad Sci U S A , 115 :E1437 , 2018 |
| Abstract : Ross_2018_Proc.Natl.Acad.Sci.U.S.A_115_E1437 |
| ESTHER : Ross_2018_Proc.Natl.Acad.Sci.U.S.A_115_E1437 |
| PubMedSearch : Ross_2018_Proc.Natl.Acad.Sci.U.S.A_115_E1437 |
| PubMedID: 29382749 |
| Gene_locus related to this paper: human-DPP8 , human-DPP9 |
| Title : The SUMO1-E67 Interacting Loop Peptide Is an Allosteric Inhibitor of the Dipeptidyl Peptidases 8 and 9 - Pilla_2013_J.Biol.Chem_288_32787 |
| Author(s) : Pilla E , Kilisch M , Lenz C , Urlaub H , Geiss-Friedlander R |
| Ref : Journal of Biological Chemistry , 288 :32787 , 2013 |
| Abstract : Pilla_2013_J.Biol.Chem_288_32787 |
| ESTHER : Pilla_2013_J.Biol.Chem_288_32787 |
| PubMedSearch : Pilla_2013_J.Biol.Chem_288_32787 |
| PubMedID: 24072711 |
| Gene_locus related to this paper: human-DPP8 , human-DPP9 |
| Title : A Novel SUMO1-specific Interacting Motif in Dipeptidyl Peptidase 9 (DPP9) That Is Important for Enzymatic Regulation - Pilla_2012_J.Biol.Chem_287_44320 |
| Author(s) : Pilla E , Moller U , Sauer G , Mattiroli F , Melchior F , Geiss-Friedlander R |
| Ref : Journal of Biological Chemistry , 287 :44320 , 2012 |
| Abstract : Pilla_2012_J.Biol.Chem_287_44320 |
| ESTHER : Pilla_2012_J.Biol.Chem_287_44320 |
| PubMedSearch : Pilla_2012_J.Biol.Chem_287_44320 |
| PubMedID: 23152501 |
| Gene_locus related to this paper: human-DPP9 |