Mocap
General
Type : Organophosphate,Insecticide,Sulfur Compound
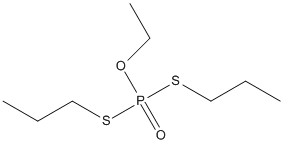
Chemical_Nomenclature : O-Ethyl-S,S-dipropylphosphorodithioate
Canonical SMILES : CCCSP(=O)(OCC)SCCC
InChI : InChI=1S\/C8H19O2PS2\/c1-4-7-12-11(9,10-6-3)13-8-5-2\/h4-8H2,1-3H3
InChIKey : VJYFKVYYMZPMAB-UHFFFAOYSA-N
Other name(s) : Ethoprop,Ethoprophos,Prophos,Rovokil,Jolt,Phosethoprop,Phophos,Holdem JOLT,MOCAP 10G
MW : 242.3
Formula : C8H19O2PS2
CAS_number : 13194-48-4
PubChem : 3289
UniChem : VJYFKVYYMZPMAB-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
References (3)
| Title : Acute toxicity and cholinesterase inhibition of the nematicide ethoprophos in larvae of gar Atractosteus tropicus (Semionotiformes: Lepisosteidae) - Mena_2012_Rev.Biol.Trop_60_361 |
| Author(s) : Mena Torres F , Pfennig S , Arias Andres Mde J , Marquez-Couturier G , Sevilla A , Protti CM |
| Ref : Rev Biol Trop , 60 :361 , 2012 |
| Abstract : Mena_2012_Rev.Biol.Trop_60_361 |
| ESTHER : Mena_2012_Rev.Biol.Trop_60_361 |
| PubMedSearch : Mena_2012_Rev.Biol.Trop_60_361 |
| PubMedID: 22458230 |
| Title : Factors influencing the ability of Pseudomonas putida strains epI and II to degrade the organophosphate ethoprophos - Karpouzas_2000_J.Appl.Microbiol_89_40 |
| Author(s) : Karpouzas DG , Walker A |
| Ref : J Appl Microbiol , 89 :40 , 2000 |
| Abstract : Karpouzas_2000_J.Appl.Microbiol_89_40 |
| ESTHER : Karpouzas_2000_J.Appl.Microbiol_89_40 |
| PubMedSearch : Karpouzas_2000_J.Appl.Microbiol_89_40 |
| PubMedID: 10945777 |
| Title : Selective, solid-matrix dispersion extraction of organophosphate pesticide residues from milk - Di Muccio_1996_J.Chromatogr.A_754_497 |
| Author(s) : Di Muccio A , Pelosi P , Camoni I , Attard Barbini D , Dommarco R , Generali T , Ausili A |
| Ref : Journal of Chromatography A , 754 :497 , 1996 |
| Abstract : Di Muccio_1996_J.Chromatogr.A_754_497 |
| ESTHER : Di Muccio_1996_J.Chromatogr.A_754_497 |
| PubMedSearch : Di Muccio_1996_J.Chromatogr.A_754_497 |
| PubMedID: 8997741 |