MUP
the chloride derivative is used in inhibition studies MUCP
General
Type : Organophosphate
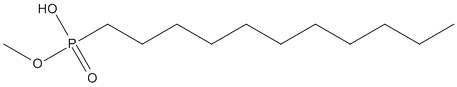
Chemical_Nomenclature : Methoxyundecylphosphonic acid
Canonical SMILES : CCCCCCCCCCCP(=O)(O)OC
InChI : InChI=1S\/C12H27O3P\/c1-3-4-5-6-7-8-9-10-11-12-16(13,14)15-2\/h3-12H2,1-2H3,(H,13,14)
InChIKey : JBVUSHKPEBKWQP-UHFFFAOYSA-N
Other name(s) : AC1L9KBG,Methoxy(undecyl)phosphinic acid,Methoxyundecylphosphinic acid,SCHEMBL9079355,DB08222,MUCP,MUPA
MW : 250.314
Formula : C12H27O3P
CAS_number :
PubChem : 446977
UniChem : JBVUSHKPEBKWQP-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : MUP ligand of proteins in family: Pancreatic_lipase || Cutinase
Stucture :
Protein : human-PNLIP || fusso-cutas
References (3)
| Title : Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands - Hodneland_2002_Proc.Natl.Acad.Sci.U.S.A_99_5048 |
| Author(s) : Hodneland CD , Lee YS , Min DH , Mrksich M |
| Ref : Proceedings of the National Academy of Sciences of the United States of America , 99 :5048 , 2002 |
| Abstract : Hodneland_2002_Proc.Natl.Acad.Sci.U.S.A_99_5048 |
| ESTHER : Hodneland_2002_Proc.Natl.Acad.Sci.U.S.A_99_5048 |
| PubMedSearch : Hodneland_2002_Proc.Natl.Acad.Sci.U.S.A_99_5048 |
| PubMedID: 11959956 |
| Title : The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate - Egloff_1995_Biochemistry_34_2751 |
| Author(s) : Egloff MP , Marguet F , Buono G , Verger R , Cambillau C , van Tilbeurgh H |
| Ref : Biochemistry , 34 :2751 , 1995 |
| Abstract : Egloff_1995_Biochemistry_34_2751 |
| ESTHER : Egloff_1995_Biochemistry_34_2751 |
| PubMedSearch : Egloff_1995_Biochemistry_34_2751 |
| PubMedID: 7893686 |
| Gene_locus related to this paper: human-PNLIP |
| Title : Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography - van Tilbeurgh_1993_Nature_362_814 |
| Author(s) : van Tilbeurgh H , Egloff MP , Martinez C , Rugani N , Verger R , Cambillau C |
| Ref : Nature , 362 :814 , 1993 |
| Abstract : van Tilbeurgh_1993_Nature_362_814 |
| ESTHER : van Tilbeurgh_1993_Nature_362_814 |
| PubMedSearch : van Tilbeurgh_1993_Nature_362_814 |
| PubMedID: 8479519 |
| Gene_locus related to this paper: human-PNLIP |