F4P
Monoacylglycerol lipase (human-MGLL) inhibitor
General
Type : Piperazine,Triazol
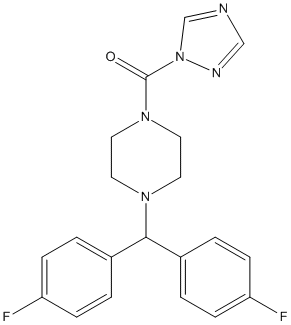
Chemical_Nomenclature : [4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-(1,2,4-triazol-1-yl)methanone
Canonical SMILES : C1CN(CCN1C(C2=CC=C(C=C2)F)C3=CC=C(C=C3)F)C(=O)N4C=NC=N4
InChI : InChI=1S\/C20H19F2N5O\/c21-17-5-1-15(2-6-17)19(16-3-7-18(22)8-4-16)25-9-11-26(12-10-25)20(28)27-14-23-13-24-27\/h1-8,13-14,19H,9-12H2
InChIKey : UIOVHJYJUUSVIG-UHFFFAOYSA-N
Other name(s) : SAR629
MW : 383.39
Formula : C20H19F2N5O
CAS_number :
PubChem : 44593853
UniChem : UIOVHJYJUUSVIG-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : F4P ligand of proteins in family: Monoglyceridelipase_lysophospholip
Stucture : 3JWE Crystal structure of human Mono glyceride lipase in complex with SAR629
Protein : human-MGLL
References (3)
| Title : Mechanistic Modeling of Monoglyceride Lipase Covalent Modification Elucidates the Role of Leaving Group Expulsion and Discriminates Inhibitors with High and Low Potency - Galvani_2022_J.Chem.Inf.Model__ |
| Author(s) : Galvani F , Scalvini L , Rivara S , Lodola A , Mor M |
| Ref : J Chem Inf Model , : , 2022 |
| Abstract : Galvani_2022_J.Chem.Inf.Model__ |
| ESTHER : Galvani_2022_J.Chem.Inf.Model__ |
| PubMedSearch : Galvani_2022_J.Chem.Inf.Model__ |
| PubMedID: 35580195 |
| Gene_locus related to this paper: human-MGLL |
| Title : Monoglyceride lipase: Structure and inhibitors - Scalvini_2016_Chem.Phys.Lipids_197_13 |
| Author(s) : Scalvini L , Piomelli D , Mor M |
| Ref : Chemistry & Physic of Lipids , 197 :13 , 2016 |
| Abstract : Scalvini_2016_Chem.Phys.Lipids_197_13 |
| ESTHER : Scalvini_2016_Chem.Phys.Lipids_197_13 |
| PubMedSearch : Scalvini_2016_Chem.Phys.Lipids_197_13 |
| PubMedID: 26216043 |
| Title : Structural basis for human monoglyceride lipase inhibition - Bertrand_2010_J.Mol.Biol_396_663 |
| Author(s) : Bertrand T , Auge F , Houtmann J , Rak A , Vallee F , Mikol V , Berne PF , Michot N , Cheuret D , Hoornaert C , Mathieu M |
| Ref : Journal of Molecular Biology , 396 :663 , 2010 |
| Abstract : Bertrand_2010_J.Mol.Biol_396_663 |
| ESTHER : Bertrand_2010_J.Mol.Biol_396_663 |
| PubMedSearch : Bertrand_2010_J.Mol.Biol_396_663 |
| PubMedID: 19962385 |
| Gene_locus related to this paper: human-MGLL |