DEUP
Diethylumbelliferyl phosphate is a selective, potent cholesterol esterase inhibitor. It blocks steroidogenesis primarily by preventing cholesterol transport into the mitochondria of steroidogenic cells. Inhibitors of cholesterol esterase are anticipated to limit the absorption of dietary cholesterol. IC50 = 11.6 microM. Substrate for testing OP-hydrolase activity (in particular the modified carboxylesterase E3 from Lucilia cuprina).
General
Type : Organophosphate,Coumarin
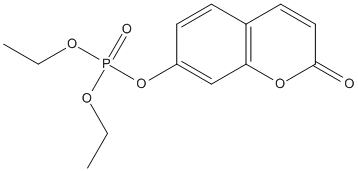
Chemical_Nomenclature : diethyl (2-oxochromen-7-yl) phosphate
Canonical SMILES : CCOP(=O)(OCC)OC1=CC2=C(C=C1)C=CC(=O)O2
InChI : InChI=1S\/C13H15O6P\/c1-3-16-20(15,17-4-2)19-11-7-5-10-6-8-13(14)18-12(10)9-11\/h5-9H,3-4H2,1-2H3
InChIKey : LZXQXHKDCNQUOF-UHFFFAOYSA-N
Other name(s) : diethyl 7-hydroxycoumaryl phosphate,O,O-diethyl O-(2-oxo-2H-chromen-7-yl) phosphate,CHEMBL1719220,DEUP,dECP,UBP,Diethylumbelliferyl phosphate,D7692_SIGMA,SCHEMBL404341
MW : 298.23
Formula : C13H15O6P
CAS_number : 897-83-6
PubChem : 16219277
UniChem : LZXQXHKDCNQUOF-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : DEUP ligand of proteins in family: Cholesterol_esterase || Hormone-sensitive_lipase_like || Arylacetamide_deacetylase || Carb_B_Arthropoda
Stucture :
Protein : human-NCEH1 || luccu-E3aest7
References (7)
| Title : Evolution of Protein Quaternary Structure in Response to Selective Pressure for Increased Thermostability - Fraser_2016_J.Mol.Biol_428_2359 |
| Author(s) : Fraser NJ , Liu JW , Mabbitt PD , Correy GJ , Coppin CW , Lethier M , Perugini MA , Murphy JM , Oakeshott JG , Weik M , Jackson CJ |
| Ref : Journal of Molecular Biology , 428 :2359 , 2016 |
| Abstract : Fraser_2016_J.Mol.Biol_428_2359 |
| ESTHER : Fraser_2016_J.Mol.Biol_428_2359 |
| PubMedSearch : Fraser_2016_J.Mol.Biol_428_2359 |
| PubMedID: 27016206 |
| Gene_locus related to this paper: luccu-E3aest7 |
| Title : Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography - Correy_2016_Structure_24_977 |
| Author(s) : Correy GJ , Carr PD , Meirelles T , Mabbitt PD , Fraser NJ , Weik M , Jackson CJ |
| Ref : Structure , 24 :977 , 2016 |
| Abstract : Correy_2016_Structure_24_977 |
| ESTHER : Correy_2016_Structure_24_977 |
| PubMedSearch : Correy_2016_Structure_24_977 |
| PubMedID: 27210287 |
| Gene_locus related to this paper: luccu-E3aest7 |
| Title : Structure and function of an insect alpha-carboxylesterase (alphaEsterase7) associated with insecticide resistance - Jackson_2013_Proc.Natl.Acad.Sci.U.S.A_110_10177 |
| Author(s) : Jackson CJ , Liu JW , Carr PD , Younus F , Coppin C , Meirelles T , Lethier M , Pandey G , Ollis DL , Russell RJ , Weik M , Oakeshott JG |
| Ref : Proc Natl Acad Sci U S A , 110 :10177 , 2013 |
| Abstract : Jackson_2013_Proc.Natl.Acad.Sci.U.S.A_110_10177 |
| ESTHER : Jackson_2013_Proc.Natl.Acad.Sci.U.S.A_110_10177 |
| PubMedSearch : Jackson_2013_Proc.Natl.Acad.Sci.U.S.A_110_10177 |
| PubMedID: 23733941 |
| Gene_locus related to this paper: luccu-E3aest7 |
| Title : Hydrolysis of organophosphorus insecticides by in vitro modified carboxylesterase E3 from Lucilia cuprina - Heidari_2004_Insect.Biochem.Mol.Biol_34_353 |
| Author(s) : Heidari R , Devonshire AL , Campbell BE , Bell KL , Dorrian SJ , Oakeshott JG , Russell RJ |
| Ref : Insect Biochemistry & Molecular Biology , 34 :353 , 2004 |
| Abstract : Heidari_2004_Insect.Biochem.Mol.Biol_34_353 |
| ESTHER : Heidari_2004_Insect.Biochem.Mol.Biol_34_353 |
| PubMedSearch : Heidari_2004_Insect.Biochem.Mol.Biol_34_353 |
| PubMedID: 15041019 |
| Gene_locus related to this paper: luccu-E3aest7 |
| Title : Kinetic efficiency of mutant carboxylesterases implicated in organophosphate insecticide resistance - Devonshire_2003_Pestic.Biochem.Physiol_76_1 |
| Author(s) : Devonshire AL , Heidari R , Bell KL , Campbell PM , Campbell BE , Odgers WA , Oakeshott JG , Russell RJ |
| Ref : Pesticide Biochemistry and Physiology , 76 :1 , 2003 |
| Abstract : Devonshire_2003_Pestic.Biochem.Physiol_76_1 |
| ESTHER : Devonshire_2003_Pestic.Biochem.Physiol_76_1 |
| PubMedSearch : Devonshire_2003_Pestic.Biochem.Physiol_76_1 |
| PubMedID: |
| Gene_locus related to this paper: luccu-E3aest7 , musdo-EST23aes07 |
| Title : Diethylumbelliferyl phosphate inhibits steroidogenesis by interfering with a long-lived factor acting between protein kinase A activation and induction of the steroidogenic acute regulatory protein (StAR) - Choi_1995_Eur.J.Biochem_234_680 |
| Author(s) : Choi YS , Stocco DM , Freeman DA |
| Ref : European Journal of Biochemistry , 234 :680 , 1995 |
| Abstract : Choi_1995_Eur.J.Biochem_234_680 |
| ESTHER : Choi_1995_Eur.J.Biochem_234_680 |
| PubMedSearch : Choi_1995_Eur.J.Biochem_234_680 |
| PubMedID: 8536719 |
| Title : Evidence that a neutral cholesteryl ester hydrolase is responsible for the extralysosomal hydrolysis of high-density lipoprotein cholesteryl ester in rat hepatoma cells (Fu5AH) - Delamatre_1993_J.Cell.Physiol_157_164 |
| Author(s) : Delamatre JG , Carter RM , Hornick CA |
| Ref : Journal of Cellular Physiology , 157 :164 , 1993 |
| Abstract : Delamatre_1993_J.Cell.Physiol_157_164 |
| ESTHER : Delamatre_1993_J.Cell.Physiol_157_164 |
| PubMedSearch : Delamatre_1993_J.Cell.Physiol_157_164 |
| PubMedID: 8408234 |