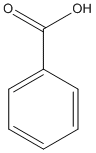
Benzoic-acid
Product of hydolysis of HODA-6-phenyl (phenyl-HODA) by Carbon-carbon_bond_hydrolase psefl-cumD. Product of hydolysis of 4-(4-nitrophenoxy)carbonylbenzoic acid (MpNPT) by idesa-mheth
General
Type : Benzoate
Chemical_Nomenclature : benzoic acid
Canonical SMILES : C1=CC=C(C=C1)C(=O)O
InChI : InChI=1S\/C7H6O2\/c8-7(9)6-4-2-1-3-5-6\/h1-5H,(H,8,9)
InChIKey : WPYMKLBDIGXBTP-UHFFFAOYSA-N
Other name(s) : Benzoate,Benzoic acid,Benzenecarboxylic acid,Dracylic acid,Carboxybenzene,Benzeneformic acid,C6H5COOH
MW : 122.12
Formula : C7H6O2
CAS_number : 65-85-0
PubChem : 243
UniChem : WPYMKLBDIGXBTP-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : Benzoic-acid ligand of proteins in family: Carboxypeptidase_S10 || Haloperoxidase || Carbon-carbon_bond_hydrolase || Cocaine_esterase || Haloalkane_dehalogenase-HLD2 || Tannase || Hydroxynitrile_lyase
Stucture :
Protein : sorbi-hnl || strau-cpoT || psefl-cumD || rhosm-cocE || rhoso-halo1 || idesa-mheth || hevbr-hnl
References (8)
| Title : Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase - Chen_2021_Angew.Chem.Int.Ed.Engl_60_21959 |
| Author(s) : Chen X , Deng X , Zhang Y , Wu Y , Yang K , Li Q , Wang J , Yao W , Tong J , Xie T , Hou S , Yao J |
| Ref : Angew Chem Int Ed Engl , 60 :21959 , 2021 |
| Abstract : Chen_2021_Angew.Chem.Int.Ed.Engl_60_21959 |
| ESTHER : Chen_2021_Angew.Chem.Int.Ed.Engl_60_21959 |
| PubMedSearch : Chen_2021_Angew.Chem.Int.Ed.Engl_60_21959 |
| PubMedID: 34351032 |
| Gene_locus related to this paper: rhosm-cocE |
| Title : Characterization and engineering of a two-enzyme system for plastics depolymerization - Knott_2020_Proc.Natl.Acad.Sci.U.S.A_117_25476 |
| Author(s) : Knott BC , Erickson E , Allen MD , Gado JE , Graham R , Kearns FL , Pardo I , Topuzlu E , Anderson JJ , Austin HP , Dominick G , Johnson CW , Rorrer NA , Szostkiewicz CJ , Copie V , Payne CM , Woodcock HL , Donohoe BS , Beckham GT , McGeehan JE |
| Ref : Proc Natl Acad Sci U S A , 117 :25476 , 2020 |
| Abstract : Knott_2020_Proc.Natl.Acad.Sci.U.S.A_117_25476 |
| ESTHER : Knott_2020_Proc.Natl.Acad.Sci.U.S.A_117_25476 |
| PubMedSearch : Knott_2020_Proc.Natl.Acad.Sci.U.S.A_117_25476 |
| PubMedID: 32989159 |
| Gene_locus related to this paper: idesa-mheth |
| Title : Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel - Koudelakova_2013_Angew.Chem.Int.Ed.Engl_52_1959 |
| Author(s) : Koudelakova T , Chaloupkova R , Brezovsky J , Prokop Z , Sebestova E , Hesseler M , Khabiri M , Plevaka M , Kulik D , Kuta Smatanova I , Rezacova P , Ettrich R , Bornscheuer UT , Damborsky J |
| Ref : Angew Chem Int Ed Engl , 52 :1959 , 2013 |
| Abstract : Koudelakova_2013_Angew.Chem.Int.Ed.Engl_52_1959 |
| ESTHER : Koudelakova_2013_Angew.Chem.Int.Ed.Engl_52_1959 |
| PubMedSearch : Koudelakova_2013_Angew.Chem.Int.Ed.Engl_52_1959 |
| PubMedID: 23303607 |
| Gene_locus related to this paper: rhoso-halo1 |
| Title : A series of crystal structures of a meta-cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) complexed with various cleavage products - Fushinobu_2005_Biosci.Biotechnol.Biochem_69_491 |
| Author(s) : Fushinobu S , Jun SY , Hidaka M , Nojiri H , Yamane H , Shoun H , Omori T , Wakagi T |
| Ref : Biosci Biotechnol Biochem , 69 :491 , 2005 |
| Abstract : Fushinobu_2005_Biosci.Biotechnol.Biochem_69_491 |
| ESTHER : Fushinobu_2005_Biosci.Biotechnol.Biochem_69_491 |
| PubMedSearch : Fushinobu_2005_Biosci.Biotechnol.Biochem_69_491 |
| PubMedID: 15784976 |
| Gene_locus related to this paper: psefl-cumD |
| Title : Crystal structure of hydroxynitrile lyase from Sorghum bicolor in complex with the inhibitor benzoic acid: a novel cyanogenic enzyme - Lauble_2002_Biochemistry_41_12043 |
| Author(s) : Lauble H , Miehlich B , Forster S , Wajant H , Effenberger F |
| Ref : Biochemistry , 41 :12043 , 2002 |
| Abstract : Lauble_2002_Biochemistry_41_12043 |
| ESTHER : Lauble_2002_Biochemistry_41_12043 |
| PubMedSearch : Lauble_2002_Biochemistry_41_12043 |
| PubMedID: 12356304 |
| Gene_locus related to this paper: sorbi-hnl |
| Title : Biochemical characterization and structural analysis of a highly proficient cocaine esterase - Turner_2002_Biochemistry_41_12297 |
| Author(s) : Turner JM , Larsen NA , Basran A , Barbas CF, 3rd , Bruce NC , Wilson IA , Lerner RA |
| Ref : Biochemistry , 41 :12297 , 2002 |
| Abstract : Turner_2002_Biochemistry_41_12297 |
| ESTHER : Turner_2002_Biochemistry_41_12297 |
| PubMedSearch : Turner_2002_Biochemistry_41_12297 |
| PubMedID: 12369817 |
| Gene_locus related to this paper: rhosm-cocE |
| Title : Structural investigation of the cofactor-free chloroperoxidases - Hofmann_1998_J.Mol.Biol_279_889 |
| Author(s) : Hofmann B , Tolzer S , Pelletier I , Altenbuchner J , van Pee KH , Hecht HJ |
| Ref : Journal of Molecular Biology , 279 :889 , 1998 |
| Abstract : Hofmann_1998_J.Mol.Biol_279_889 |
| ESTHER : Hofmann_1998_J.Mol.Biol_279_889 |
| PubMedSearch : Hofmann_1998_J.Mol.Biol_279_889 |
| PubMedID: 9642069 |
| Gene_locus related to this paper: psefl-cpoF , strau-brpa2 , strau-cpoT , strli-cpoL |