ABL-127
ABL127 is reported to inhibit protein phosphatase methylesterase 1 (PPME1)
General
Type : Azetidin
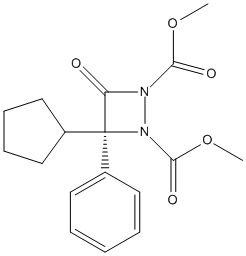
Chemical_Nomenclature : dimethyl (3R)-3-cyclopentyl-4-oxo-3-phenyldiazetidine-1,2-dicarboxylate
Canonical SMILES : COC(=O)N1C(=O)C(N1C(=O)OC)(C2CCCC2)C3=CC=CC=C3
InChI : InChI=1S\/C17H20N2O5\/c1-23-15(21)18-14(20)17(13-10-6-7-11-13,19(18)16(22)24-2)12-8-4-3-5-9-12\/h3-5,8-9,13H,6-7,10-11H2,1-2H3\/t17-\/m0\/s1
InChIKey : ZRHWCAFAIHTQKD-KRWDZBQOSA-N
Other name(s) : ABL127,ML174,SR-01000786812,SR-01000786812-5,MLS001250337
MW : 332.35
Formula : C17H20N2O5
CAS_number :
PubChem : 24856225
UniChem : ZRHWCAFAIHTQKD-KRWDZBQOSA-N
IUPHAR : 8608
Wikipedia :
Target
Families : ABL-127 ligand of proteins in family: PPase_methylesterase_euk
Stucture :
Protein : human-PPME1
References (3)
| Title : Involvement of PP2A methylation in the adipogenic differentiation of bone marrow-derived mesenchymal stem cell - Ikeda_2020_J.Biochem_168_643 |
| Author(s) : Ikeda S , Tsuji S , Ohama T , Sato K |
| Ref : J Biochem , 168 :643 , 2020 |
| Abstract : Ikeda_2020_J.Biochem_168_643 |
| ESTHER : Ikeda_2020_J.Biochem_168_643 |
| PubMedSearch : Ikeda_2020_J.Biochem_168_643 |
| PubMedID: 32663263 |
| Gene_locus related to this paper: human-PPME1 |
| Title : A high-throughput, multiplexed assay for superfamily-wide profiling of enzyme activity - Bachovchin_2014_Nat.Chem.Biol_10_656 |
| Author(s) : Bachovchin DA , Koblan LW , Wu W , Liu Y , Li Y , Zhao P , Woznica I , Shu Y , Lai JH , Poplawski SE , Kiritsy CP , Healey SE , DiMare M , Sanford DG , Munford RS , Bachovchin WW , Golub TR |
| Ref : Nat Chemical Biology , 10 :656 , 2014 |
| Abstract : Bachovchin_2014_Nat.Chem.Biol_10_656 |
| ESTHER : Bachovchin_2014_Nat.Chem.Biol_10_656 |
| PubMedSearch : Bachovchin_2014_Nat.Chem.Biol_10_656 |
| PubMedID: 24997602 |
| Gene_locus related to this paper: human-PPME1 |
| Title : Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors - Bachovchin_2011_Proc.Natl.Acad.Sci.U.S.A_108_6811 |
| Author(s) : Bachovchin DA , Mohr JT , Speers AE , Wang C , Berlin JM , Spicer TP , Fernandez-Vega V , Chase P , Hodder PS , Schurer SC , Nomura DK , Rosen H , Fu GC , Cravatt BF |
| Ref : Proc Natl Acad Sci U S A , 108 :6811 , 2011 |
| Abstract : Bachovchin_2011_Proc.Natl.Acad.Sci.U.S.A_108_6811 |
| ESTHER : Bachovchin_2011_Proc.Natl.Acad.Sci.U.S.A_108_6811 |
| PubMedSearch : Bachovchin_2011_Proc.Natl.Acad.Sci.U.S.A_108_6811 |
| PubMedID: 21398589 |
| Gene_locus related to this paper: human-PPME1 |