P-nitrophenol
General
Type : pNP, Chromogen
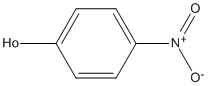
Chemical_Nomenclature : 4-nitrophenol
Canonical SMILES : C1=CC(=CC=C1[N+](=O)[O-])O
InChI : InChI=1S\/C6H5NO3\/c8-6-3-1-5(2-4-6)7(9)10\/h1-4,8H
InChIKey : BTJIUGUIPKRLHP-UHFFFAOYSA-N
Other name(s) : pNP || NPO || 4-Nitrophenol || P-nitrophenol || Phenol, 4-nitro- || Niphen || Paranitrophenol
MW : 139.11
Formula : C6H5NO3
CAS_number : 100-02-7
PubChem : 980
UniChem : BTJIUGUIPKRLHP-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : Polyesterase-lipase-cutinase, Hormone-sensitive_lipase_like
Protein : idesa-peth, 9bact-H6BDX1, 9delt-EstD11
References (2)
| Title : Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family - Miguel-Ruano_2021_Comput.Struct.Biotechnol.J_19_1214 |
| Author(s) : Miguel-Ruano V , Rivera I , Rajkovic J , Knapik K , Torrado A , Otero JM , Beneventi E , Becerra M , Sanchez-Costa M , Hidalgo A , Berenguer J , Gonzalez-Siso MI , Cruces J , Rua ML , Hermoso JA |
| Ref : Comput Struct Biotechnol J , 19 :1214 , 2021 |
| Abstract : Miguel-Ruano_2021_Comput.Struct.Biotechnol.J_19_1214 |
| ESTHER : Miguel-Ruano_2021_Comput.Struct.Biotechnol.J_19_1214 |
| PubMedSearch : Miguel-Ruano_2021_Comput.Struct.Biotechnol.J_19_1214 |
| PubMedID: 33680362 |
| Gene_locus related to this paper: 9delt-EstD11 |
| Title : Structural insights of a hormone sensitive lipase homologue Est22 - Huang_2016_Sci.Rep_6_28550 |
| Author(s) : Huang J , Huo YY , Ji R , Kuang S , Ji C , Xu XW , Li J |
| Ref : Sci Rep , 6 :28550 , 2016 |
| Abstract : Huang_2016_Sci.Rep_6_28550 |
| ESTHER : Huang_2016_Sci.Rep_6_28550 |
| PubMedSearch : Huang_2016_Sci.Rep_6_28550 |
| PubMedID: 27328716 |
| Gene_locus related to this paper: 9bact-H6BDX1 |